Open source and open science
eLife was founded on open principles, with open access at the heart of its publishing agenda. To bolster this, the nascent technology team adopted a strategy that ensured everything they built would be available as open-source software, sharing code under an OSI-approved licence on GitHub. To augment this open strategy, they also opted to use open-source tools when outsourcing or buying services and to focus on collaborating with others working in open-source software where they could.
Throughout the evolution of software products at eLife these open standards have endured, but the approach to how the team makes the software open has changed. Early attempts with products like eLife Lens and Continuum concentrated on making the codebase open and well-documented, with smaller amounts of effort invested in community building and reuse.
The team’s approach changed with the Libero Suite of products. The design philosophy was to create a ‘permissible’ rather than ‘prescribed’ user experience built on software intended for other software teams to use, extend and host themselves. While some innovative organisations were keen to collaborate on Libero, and a community around it developed, the team later moved to the current model: building open scholarly infrastructure with transparent decision-making and where reuse is part of the product itself.
You can see this difference in Sciety and the latest addition to the team’s portfolio, the Enhanced Preprint Platform that powers eLife’s new Reviewed Preprints. Both products are part of a larger ecosystem and allow multiple organisations to use the same technology through subtle branded areas and aggregation of content – all built on a codebase that is openly licenced, well documented and supported by a growing community of interested organisations.
Here we want to explore the significant products worked on at eLife over the last decade to show how the approach to openness, the feedback from the community and the changes in methods of collaboration have all contributed to the way we’re building technology for the ‘publish, review, curate’ (PRC) ecosystem today.
eLife Lens – the first open-source product for eLife
When reading articles on eLife’s website, you’ll be presented with a ‘side by side’ view as an option. This innovative way of presenting the article allows figures or references to be scrolled separately from the article's content. Readers found this useful when the text referred to figures frequently or when they explored the references in more detail. The software that provides this functionality is eLife Lens which was the first tool that eLife made open source. Given it was separate enough from the journal software, it was a great choice to share openly, as it could be easily adopted by others and adapted to different use cases.
We’ve seen this being used in similar ways to eLife but also as a way to self-host a journal, an offline article reader, and with machine-learning algorithms to display recommendations in context alongside an article. We’ve also seen this used in other journals through its inclusion as a plugin in PKP’s Open Journal System.
Continuum for automated, continuous publishing
A major release for eLife was its first open-source publishing platform that powered the journal at elifesciences.org. It was remarkable to hear staff talking about the levels of agency they now had and how this led to more innovative thinking across the publishing operation because vendors’ systems no longer constrained them. Continuum introduced continuous publication and intelligent versioning, giving eLife authors and publishers the freedom to tell their stories in full in a way that suited the work.
Continuum was also the first time that eLife was able to invest in improving the user experience and accessibility of the journal website, which was noticed by other publishers and the main driver for the growing interest we saw in others attempting to adopt the software. At the time though, the software was built specifically for eLife’s publishing workflow with all the idiosyncrasies that came with it. This made it more difficult for others to adopt than eLife Lens unless they were willing to adapt their own workflow to match.
With the growing energy around innovation at eLife, interest in our user experience design, and the acknowledgement that a monolithic software like Continuum was difficult to adopt, we decided to renew some parts of the publishing platform that would allow us to release the next version in 2017.
A new user experience for eLife in ‘eLife 2.0’
Around the time of Continuum’s initial launch, web browsing experiences were getting richer for all types of websites and people were increasingly using a variety of device form factors. Usage of mobile phones and tablets to browse the eLife journal was growing, and the default use case of downloading a PDF to read an article was becoming less popular, particularly with early-career researchers who had grown up in a web-centric world.
We felt we were in a position to offer a mobile-centric, accessibility-focused and web-based reading experience for the eLife journal that would set the standard for performance and reliability in online journals. To ensure this could be even more reusable than previous open-source products from eLife, we also changed the architecture to provide several independent services – separating the journal article rendering from the article storage and the content management from the publishing workflow. The result was the software we still use today to power eLife, which we called ‘eLife 2.0’.
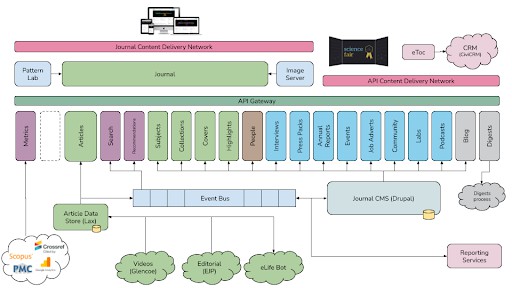
eLife 2.0 high-level architecture showing all the separately reusable components.
To further aid the reuse, we also made our design patterns – the HTML and CSS used to render the various website components – available using a pattern library so that others could easily copy and paste the parts they needed, even if they didn’t want to adopt our look and feel completely.
A significant addition to this platform in 2020 was the development of the Executable Research Article. We worked with Stencila and their technology that allows authors to present articles using interactive figures with the code and data that generates them visible and editable on the web. While immediately useful for the journal, this paves the way for authors to directly submit Python and R notebooks for peer review or post preprints in an interactive form to fully utilise the technology of the web in conveying their findings.
Open-source technology for reuse
The success of Continuum in enabling the team at eLife to innovate in their publishing efforts was contagious. The next obvious area to address was our peer review management system. We had learned lessons from making things too eLife-specific so chose to work with others from the outset.
Peer review management
The Coko Foundation had worked on their PubSweet software to power book and journal review systems. The book production platform then known as Editoria (but later renamed Ketida) offered many of the modern collaboration features we were looking for, so adapting that into a journal submission system seemed like a useful first step.
This submission system, Libero Reviewer, has been in use at eLife to collect initial submission information since 2020, and the underlying libraries we worked on with Coko since 2018 are now part of Kotahi. This modern scholarly publishing platform supports the PRC model, and is used to publish all of eLife’s public reviews and manage the peer-review process for several Sciety groups.
The original plan for Libero Reviewer (as the name suggests) was to have eLife use it to manage the entire peer-review process. It used a ‘permissive’ rather than ‘prescriptive’ approach that didn’t constrain the way the system was used, featuring permission levels and step-by-step wizards. This would have allowed the team to innovate quickly and offer a unique experience to authors, editors and reviewers because changes or experiments to the workflow would not require significant software development time. We still intend to offer this level of innovation, but the move to a PRC model at eLife means we’ll be making a longer-term investment in Kotahi which offers the same outcomes. Therefore, Libero Reviewer is no longer being developed.
Article publishing, XML conversion and typesetting for production workflows
Part of the Libero offering was to combine Libero Reviewer with other products in the Libero Suite that offered publishing and production workflows too.
Libero Publisher was designed to be a more developer-friendly version of Continuum, enabling a group of innovative and change-tolerant publishers to switch to open-source software. We had plenty of interest in the platform, but the undertaking was simply too large for our team alone. When the move to a PRC model emerged as a more attractive option to allow these publishers to change the way they publish, we decided to focus our efforts there instead.
A similar strategy meant that we also ceased development of Libero Editor – a JATS XML editor that helps improve the production process. This was much further along with its development so we were able to pass it on to new stewards. Our previous collaboration with Coko made them an obvious choice to provide stewardship for this software. Their operating model means you can sponsor them to work on Libero Editor directly for you or contribute to help incorporate it into existing software like Kotahi.
The greatest benefit of working on the Libero Suite was the building of an open-source community that still works with us today. We had advice from the people behind the Apache Foundation and the Apereo Foundation, chose a governance model, and worked with partners at Hindawi, Digirati and Coko to form a steering committee that helped define how the product grew. All came with invaluable lessons to help us define a new operating model for our open-source software going forward.
Technology to support ‘publish, review, curate’
The biggest shift in our approach to building open-source software has been to focus on the users of the software, speaking directly with readers, authors and reviewers rather than gathering their needs through publishers. While there are a growing number of communities looking to adopt the PRC model, it is still new for many researchers so understanding how we can best serve their needs in this new era will ensure we shape our products in the most effective way possible.
We used the lessons we have learned from previous open-source endeavours and the connections we’d made with collaborators, to fully focus on creating software to support PRC that we developed, hosted and maintained but offered others to use in diverse ways. This meant we could commit to the Principles of Open Scholarly Infrastructure and offer a place for groups to practise PRC in the open, with no additional cost to them.
Exploration and curation of evaluated preprints with Sciety
The first product that enabled others to take part in PRC was Sciety. This started as a simple aggregator for peer reviews of preprints, bringing together the review output from pioneering groups like Peer Community In, Review Commons, PREreview and the Novel Coronavirus Research Compendium. When other groups joined, providing ‘highlights’ and ‘AI screenings’, we realised that PRC was much more than human peer review in the traditional sense and soon started aggregating all types of evaluation.
The promise of aggregated content quickly gained traction in the scholarly publishing community. People were excited about the potential of the next stage of Sciety’s development – providing curation facilities to groups and individuals. Beginning with a simple way to personalise your own feed by following groups and later by saving articles to your own personal list, Sciety started to allow individuals to discover content more easily. These acts of organising the preprint literature fed back into the discovery mechanisms as lists became destinations for readers.
Now, Sciety is focused on enabling individuals and groups to do more of that activity. Features that enable multiple lists with customised descriptions are being worked on right now, and ideas around annotating why a preprint is on your list are being tested with a few well-known curators. Feeding all of this information back into Sciety’s data will allow us to further organise the preprint literature and accelerate the discovery of preprints and their reviews.
The community around Sciety is, therefore, quite different to previous communities we had built. Groups like PeerRef are able to use Sciety in place of software they would have to build themselves, and groups like Biophysics Colab have been using Sciety as their home on the web to showcase their evaluation activities.
An enhanced preprint platform to support the future of scholarly communication
The latest major open-source software platform to be released by eLife is the Enhanced Preprint Platform, which powers the Reviewed Preprints recently launched as part of eLife’s move to a new PRC model that aims to offer peer review without the gatekeeping.
While the current examples are all branded with eLife elements, the platform itself can take article information from bioRxiv and medRxiv, coupled with review and assessment information from Sciety, and create an experience for anyone wanting to offer a similar PRC model. We’ll be offering this to more groups in the future.
Future technology and innovation at eLife
This latest model of building software that fosters a community of users and organisations together seems to be the one that allows us to make the broadest and most radical change in the industry.
Sciety, the Enhanced Preprint Platform and Kotahi are all pieces of a larger PRC ecosystem that includes preprint servers (like bioRxiv, medRxiv, Research Square and SciELO Preprints), reviewer-centric websites (like PREreview) and community-led publishing platforms (like PubPub) that can all grow by working together. Different combinations of these technologies allow many flavours of PRC to come to fruition, supporting everything from journal clubs to open research platforms from funders and institutions.
Head of Technology and Innovation, Paul Shannon, says. “We plan to continue building these innovative tools and growing our collaborations. I’ve only been here for 6 of the 10 years that eLife has been on this journey, but I can see how we continue to listen and learn from people that care about open science. I’m pleased we can support that with adaptable and robust open-source software that genuinely benefits researchers.”
This blog is part of a special collection celebrating 10 years of eLife.
#
We welcome comments/questions from researchers as well as other journals. Please annotate publicly on the article or contact us at hello [at] elifesciences [dot] org.
For the latest in published research sign up for our bi-weekly email alerts. You can also follow @eLife on Twitter.




