By Tyler Whitehouse, Dav Clark and Emmy Tsang
Computational reproducibility should be trivial but it is not. Though code and data are increasingly shared, the community has realised that many other factors affect reproducibility, a typical example of which is the difficulty in reconstructing a work’s original library dependencies and software versions. The required level of detail documenting such aspects scales with the complexity of the problem, making the creation of user-friendly solutions very challenging.
The need for such solutions is something that authors, journals and readers have long agreed on, but despite various efforts there hasn’t been much progress until now. Over the last two years, new approaches focusing on optimizing the individual user experience have shown that fairly robust reproducibility is possible for a broad spectrum of users without a whole lot of effort or skill by anybody but the software developers.
The new solutions leverage software containers and follow a familiar pattern of standardization, simplification and automation to decrease mistakes and confusion, thereby improving the “user-friendliness” and robustness of computational reproducibility. The pivot towards opinionated frameworks and browser-based tools has opened up a path that to some extent obviates the “hard to teach and easy to get wrong” manual techniques of previous approaches.
Despite these improvements, the road to making computational reproducibility the norm is still a long one. In addition to continuing technical difficulties, the practical conception of reproducibility continues to evolve along with emerging technologies. Now that the underlying technology has significantly changed, it is clear that the practice of reproducibility itself also needs an update.
Over the last year, the team at Gigantum has evaluated the reproducibility of various journal articles, and this experience has helped us formulate some basic questions to keep us focused on what is crucial for developing a useful platform. These questions are:
- Why should anybody care about reproducibility?
- What separates difficult from easy reproducibility problems?
- How should we evaluate a reproducibility solution?
- What is the role of re-execution in verification and reproducibility?
- What is the role of certification in verification and reproducibility?
Far-ranging discussions between publishers and researchers have reinforced the relevance of these questions and the variety of perspectives on their answers. In particular, eLife has been a vocal participant in discussions on computational reproducibility and the tools that help make it a reality. For example, various solutions like Binder, Stencila and Gigantum have been announced in Labs posts over the past two years. So, another Labs post seemed the proper place to put these questions to the broader community and share our perspective.
Asking and answering these questions will hopefully initiate a new discussion around improving the quality and reproducibility of research, the user experience of peer review, and the practical and economic realities of computational reproducibility.
Why should anybody care?
An individual’s motivations around computational reproducibility determine their standards and approach, but regardless of the definition (see a good one in The Turing Way), reproducibility is an important proxy for something much more difficult: replicability. The centrality of replicability to the scientific method connects reproducibility to the upward spiral of knowledge and the common good. More practically, reproducibility is a way to provide quality assurance in peer review with the efficiency needed for the massive volume of scientific knowledge flooding through the publishing infrastructure.
Despite their purity and practicality, these motivations don’t seem sufficient to make reproducibility an overriding and actionable concern to most researchers and publishers. To motivate the average researcher or publisher to care about reproducibility, we need more human motives: ease and credibility. While credibility probably doesn’t need a definition, by ease we mean the time, skill and money required to do something.
Reproducibility for verification is a point of contact between the reputations of scientists and the journals that publish their work. Publishers’ reproducibility requirements make it easier for reviewers to scrutinize an author’s work, but they also help to preserve the journal and authors’ reputations as sources of valid and worthwhile science.
Reproducibility is also a point of contact between the authors and their peers. Community-based demands for reproducibility make it easier to subject the author’s work (and the journal’s review process) to knowledgeable and competitive scrutiny, significantly reducing the difficulty and time required to understand and then accept or challenge the results.
Prioritizing ease of interrogation and reuse in computational reproducibility preferences a dynamic type of reproducibility that merges into replicability, wherein the techniques producing a specific result are intelligible, re-executable and extendable to novel data or computational techniques. Since replication is the ultimate aim, we should all be prioritizing ease of interrogation and reuse in computational reproducibility.
The practical conception of reproducibility continues to evolve along with emerging technologies.
What Separates Difficult From Easy?
Defining ease as minimizing the amount of time, skill and money is reasonable, but it depends on the computation and whoever is trying to reproduce it. Since the target audience varies, it is productive to model why computations are intrinsically difficult to reproduce.
One toy model breaks a workflow into an expensive first computational step to create outputs, followed by a second step of lighter-weight analysis of those same outputs. In the figure below, Input Data and Main Computations comprise the first step, and the Intermediate Data and Analysis comprise the second step. For example, training a deep-learning or random forest classifier and analyzing its effectiveness on test data fits into this model. Many bioinformatics workflows fit into this model as well.
In the figure, the line indicates where the majority of time, skill and money would be spent to reproduce computations. Reproducing computations on the left-hand side is “difficult” while reproducing those on the right-hand side is “easy”, i.e. there are existing solutions that significantly reduce the time, skill and money to reproduce them.
Grappling with these situations doesn’t just reduce to reproducing software environments. Large data, specialized hardware and long compute times make such workflows difficult or infeasible to reproduce. Any framework that wants to open up the processes left of the line must account for access to potentially massive data and the implementation of expensive and highly technical computing infrastructure. This always costs time, skill and money no matter who is doing it.
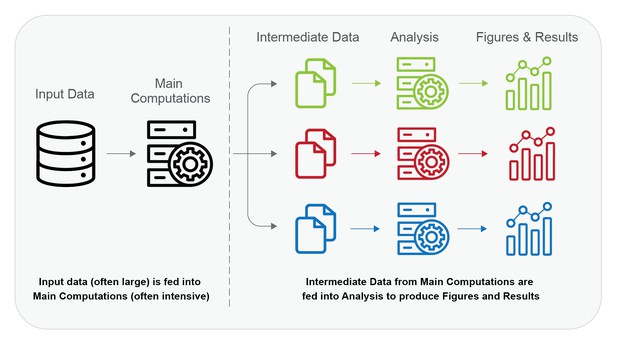
The dotted line is a practical division between computations that are difficult to reproduce and those that are easy or feasible in this toy model.
While not perfect, this toy model covers many cases and lets us set expectations and choose approaches. A more nuanced model would account for the difficulties that crop up in the right-hand side because the Intermediate Data and Analysis are not all necessarily lightweight. Models of this type can be used to determine which parts of a workflow are reasonable to be made re-executable for verification, and which parts are better verified through methods other than strict re-execution.
A Rubric to Evaluate Reproducibility Solutions
We also need models to help evaluate reproducibility solutions themselves. We propose a fairly effective one below. The terms are overloaded and somewhat simplistic, but they reveal important aspects of the story.
- The Producer: This is whoever created the computation to be reproduced. For example, it could be a graduate student who wrote a paper with accompanying code and data.
- The Project: This is what is to be reproduced, i.e. the materials (code, environment, data and instructions) and the procedure needed to reproduce the result. For example, it could be a GitHub repository of code, data, a requirements.txt file, and a README file.
- The Infrastructure: This is the hardware and data storage (actual or virtual) required to run a Project, i.e. where the computation or reproduction happens. It can also include software. For example, it could be a Ubuntu 16.04 workstation with an Nvidia GPU, or a cloud server with fast and free access to data in Amazon S3.
- The Consumer: This is whoever will reproduce the results of the Project or adapt the Project for a novel question. In the first case, it could be a reviewer for a journal article, or a testing and evaluation team from a government lab. In the second, it could be a researcher, policy analyst, or an interested amateur.
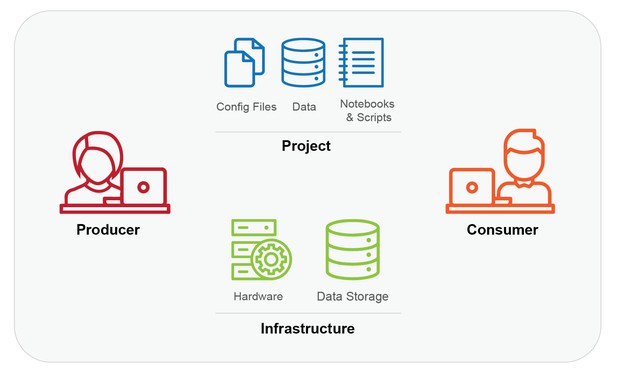
The four parts of the basic model. Producers create the Project to work on a given Infrastructure, and Consumers need access to both the Project and an appropriate Infrastructure to reproduce the work (People icons used under creative commons license from lastspark of the Noun Project).
These simple terms let us make the following very clear statement:
“Producers should be able to create a Project that can be reproduced by a Consumer on available Infrastructure.”
A solution’s value is in its ease for Producers and Consumers, how well it allows reproduction, and its ability to conform to available Infrastructure. We evaluate using the following:
- How easy is it for the Producer to create a reproducible Project?
- How easy is it for the Consumer to reproduce the Project?
- How flexible is the Infrastructure in terms of location, configuration and control?
- How much does the Consumer need to re-execute to verify that things work?
- How easy is it for the Project to be reused or repurposed by the Consumer?
Evaluation using these criteria is subjective, and independent observers can easily disagree due to different skills, available time and funds. Despite this, the relationships between them are less subjective. Producers, Projects and Infrastructure are tied together in ways that can quantifiably either promote or inhibit reproduction, depending on the end goal. Evaluating trade-offs in these relationships helps to fit an approach to a task.
Why these specific criteria?
These questions are fundamental for a few reasons. First, the adoption of techniques by Producers is a major bottleneck in reproducibility. Solutions that make reproducibility easy are crucial because difficult methods have a high barrier to adoption. Solutions that are accessible or reduce total effort will be adopted even by Producers that don’t care about reproducibility per se, thus improving reproducibility for all stakeholders.
A major consideration in questions of reproducibility is the position and time of the Consumer. Failures in reproducibility often hinge on technical asymmetries between Producers and Consumers, and the work required by a Consumer affects their inclination to reproduce or verify something. Ease for the Consumer promotes validation and diffusion of a result.
Another consideration is choice in the location, configuration and control of the needed Infrastructure. A simple analysis on a csv file is very different than large-scale machine learning, and the dependence on Infrastructure requires the right sizing of hardware and costs depending on the Consumer’s needs and abilities.
Concurrent with minimizing effort by the Consumer is that of minimizing effort by the computer. Solutions that employ certification (i.e. a complete and comprehensive record of what was done) are better for minimizing the effort and costs of verification than redundant soup-to-nuts re-execution.
Finally, as we’ve argued before, simple re-execution shouldn’t be the end goal. Interrogation, reuse and incorporation are more in line with the goals of the scientific community. Code and data should be easily discoverable, reusable and extendable in ways that the Consumer can own and maintain themselves.
Interrogation, reuse and incorporation are more in line with the goals of the scientific community.
Re-evaluating Re-execution
Consider reproducing a “difficult” Project that uses a GPU. This necessitates a specific and costly Infrastructure, and though software plays a role, the hardware is ultimately the most significant resource bottleneck. Container-based approaches capture environments but provisioning and managing the necessary Infrastructure can’t be reduced to a Dockerfile.
Due to this and other difficulties, Projects that are reproducible on their face (because the necessary data, code and software environments are provided) can actually be out of reach.
Considering the difficulties of re-executing computationally heavy projects, it seems fair to ask if re-execution should be avoided. The time and money spent reproducing computations are daunting, and the related concerns increase when considering environmental impacts. While everybody knows that bitcoin mining is terrible for the environment, it is only recently that the environmental consequences of large-scale machine learning are being investigated. They aren’t great.
There should be a discussion about the value of re-execution in the context reproducibility. Why is it even needed?
Questioning the primacy of re-execution isn’t just about value or necessity. It is also a question of access, because if re-execution remains the primary means of verification then validating large computations will be off-limits to those without enough cash, potentially leading to a “reproducibility gap”, wherein only those with sufficient money can play.
If you compare the budget of a pharmaceutical company to the budget of a university researcher, this type of looming asymmetry should give you serious pause. Furthermore, science already has a structural problem with unequal access across the globe and massive paywalls. Pinning verification to financial resources seems like a bad way to democratize research.
After spending the last two years developing a reproducibility platform that makes re-execution trivial to do, the team at Gigantum doesn’t say any of this lightly.
Due to this and other difficulties, Projects that are reproducible on their face can actually be out of reach.
Certification as a Means and an End
As science has become increasingly computational, you would think that the certification of digital processes would be a solved problem. As any researcher will tell you, it isn’t. Proper and trustworthy documentation of the digital process is difficult for technical and human reasons.
Until now, the kinds of practices that the Carpentries teach were the best path to the robustness of use and documentation needed to overcome the techno-human frailties that lead to failures in reproducibility. However, manual approaches don’t capture what happens upon execution, and version control is not an article of record for computation. Re-execution fills this gap but the fact remains that it is essentially a proxy for certification.
When we step back and look at current approaches to reproducibility, it seems clear that treating re-execution as the gold standard has more or less defined today’s most popular approaches to the problem.
Work Environments, Not Reproducibility Platforms
We believe that taking a post-hoc approach to reproducibility is the wrong way to do things. In our experience, frameworks that provide retrofitting for reproducibility are really just complicated systems to re-execute archives. This forces any sort of certification to happen within the post-hoc process, incurring needless re-execution and other difficulties.
If computational activity is recorded as it is created, then you have the beginnings of certification. This is why we’ve been advocating for work environments that provide a record of what computations were done, thus eliminating the need to re-execute.
The difficulty lies in making sure that the certification creates complete trust and captures all relevant aspects of the process. In Gigantum we use automated git versioning and metadata to capture and document computational work done in Jupyter or RStudio. It isn’t quite certification because someone sufficiently skilled and malicious might be able to alter those records, but it is close.
We think that certification as both a means and an end for verification is the best way to avoid the quandaries of re-execution. It is a reasonable path to satisfy the rigour of scientific review with the required economy and speed.
An Invitation To Improve Science
The above is a retrospective summary of the explorations and advances we’ve made in developing the MIT-licensed Gigantum Client. The evaluation rubric represents what we’ve prioritized in our approach in trying to understand and tackle “difficult” reproducibility problems – moving further to the left side of our diagram where expensive (and energy-intensive) compute meets ever-increasing quantities of data. Our experience with the challenges has led us to take a path of recording activity for inspection rather than just providing the ability to re-execute.
The Gigantum “Activity Record” lays a solid foundation for certification of the exact steps taken in data analysis and reporting. Going forward, we will continue to work on minimizing the need to re-execute computational work that’s already been done, and engaging with the research community in a virtuous cycle:
- As “menial” steps are automated, researchers can focus on further improving best practices from a strong baseline as they continue to produce data.
- As best practices are refined and standardized, they can be continually streamlined and ultimately automated.
- And so researchers’ cognitive burdens around menial tasks are further reduced, and the cycle can continue.
Essential to this cycle is active conversation within a community of researchers – so we hope you will share your own best practices, whether they are done in Gigantum or another platform (including DIY). We’d also love to hear feedback on our rubric and the primary dimensions we’re describing for reproducibility. What are we missing? What have we got right?
And if you don’t know about Gigantum, you can read about it here, try it online via the big blue “demo” button on our homepage, or just tweet at @gigantumscience and ask us a question!
#
We welcome comments, questions and feedback. Please annotate publicly on the article or contact us at innovation [at] elifesciences [dot] org.
Do you have an idea or innovation to share? Send a short outline for a Labs blogpost to innovation [at] elifesciences [dot] org.
For the latest in innovation, eLife Labs and new open-source tools, sign up for our technology and innovation newsletter. You can also follow @eLifeInnovation on Twitter.




