By Aziz Khan and Tomasz Konopka
Placing research in the context of existing knowledge is a critical step in the scientific process; we routinely look for information about genes, chemicals and drugs, genetic variants, and other entities. Vast amounts of relevant data are available in databases, and their breadth and depth are growing. The ClinVar archive which collects associations between genetic variants and diseases, for example, increased in size by 60% in the past year. At the same time, smaller datasets are published on a regular basis. The knowledge landscape in the biomedical domain is changing rapidly and that is exciting, but this rapid change and growth also create challenges in how to effectively use this wealth of information in day-to-day research.
How do we currently use open biomedical data in everyday situations? Querying a search engine, of course, is a familiar route that can reveal relevant primary sources. Alternatively, it is possible to navigate to a specific data portal or data-integration platform and search on the domain-specific site. These approaches are effective, but it can nonetheless be time-consuming to browse multiple entities, find niche details, and verify that a data aggregator has up-to-date information.
Such concerns are continually discussed by data creators as well as data curators. The FAIR initiative, in particular, published guiding principles for maximising the value of open data. These provide concrete definitions for what it means for data to be findable, accessible, interoperable and reusable (FAIR). As a result of these principles, as well as independent developments in web technology and the efforts of biocuration teams, much biomedical data are today accessible from primary sources through direct channels.
The ability to download data from primary sources, whenever needed, opens exciting possibilities for data discovery and analysis. Aiming to streamline access to open data, we developed a browser extension called FAIR-biomed for previewing snippets of information from any web page. It works inside a browser to access specialist databases without the need to switch tabs, open new windows, or type URLs.
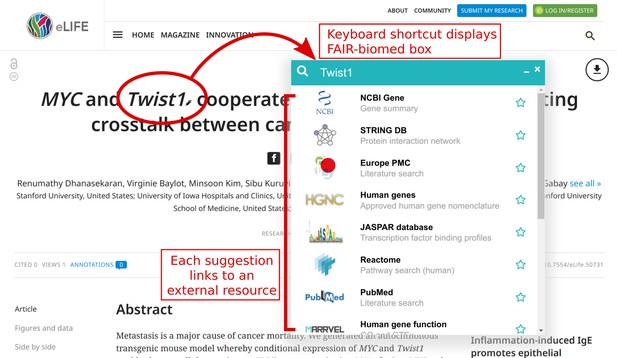
FAIR-biomed provides background information on a topic from any web page, e.g. a journal article.
The extension takes some text as input, provides a list of relevant biomedical resources, and composes queries to a chosen database. It then fetches results from that resource and displays a subset of information. Within a few keystrokes, it is possible to see a topic from several perspectives.
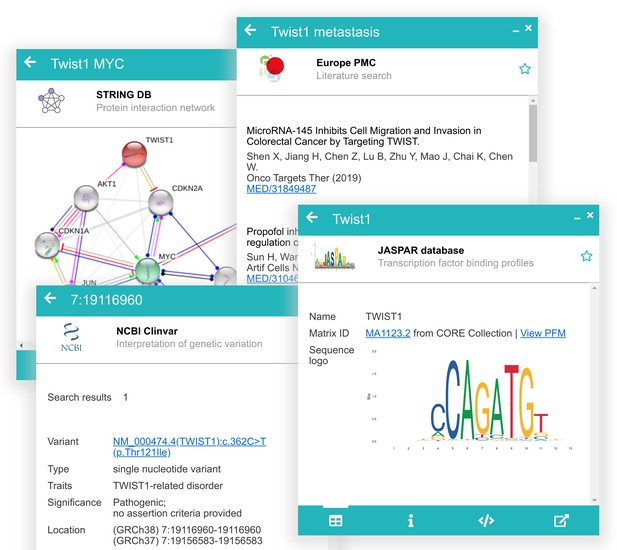
Examples of data previews showing a protein interaction network from STRING, related publications from Europe PMC, known pathogenic variants from ClinVar, and transcription factor binding profiles from JASPAR.
The idea of previewing data within web pages has a long history. For example, Reflect was an early example of a browser extension, and GIX is a more modern project. These tools focus on providing information about genes from preselected data sources. FAIR-biomed is similar in some regards, but also offers unique features and possibilities. It defers data curation to the specialised resources and only retrieves relevant portions on demand. It is also not limited to searching for gene names and can thus inform on publications, mouse models, genomic regions, variants, and other entities.
FAIR-biomed is open-source (GitHub repository) and has a modular structure. Its library comprises independent components that are each responsible for interfacing with one data resource.

The plugin library interfaces with 21 distinct resources and can be expanded organically.
This library can be extended with more components and can accept contributions from the community. Each component is defined by a logo, a description of the primary resource for a page of credits, and a javascript file. Existing components average only ~60 lines of code each and can be used as templates. If you would like to contribute to the project and add more plug-ins, please check out our project documentation.
We are also steadily adding more plugins to FAIR-biomed’s library. We invite you to try the extension out, and please do let us know if there are specific plug-ins that you would like to see in FAIR-biomed. We welcome all feedback from the research community.
Also compatible with other browsers, see installation documentation.
#
We welcome comments, questions and feedback. Please annotate publicly on the article or contact us at innovation [at] elifesciences [dot] org.
Do you have an idea or innovation to share? Send a short outline for a Labs blogpost to innovation [at] elifesciences [dot] org.
For the latest in innovation, eLife Labs and new open-source tools, sign up for our technology and innovation newsletter. You can also follow @eLifeInnovation on Twitter.




