Ovaries absent links dLsd1 to HP1a for local H3K4 demethylation required for heterochromatic gene silencing
Figures
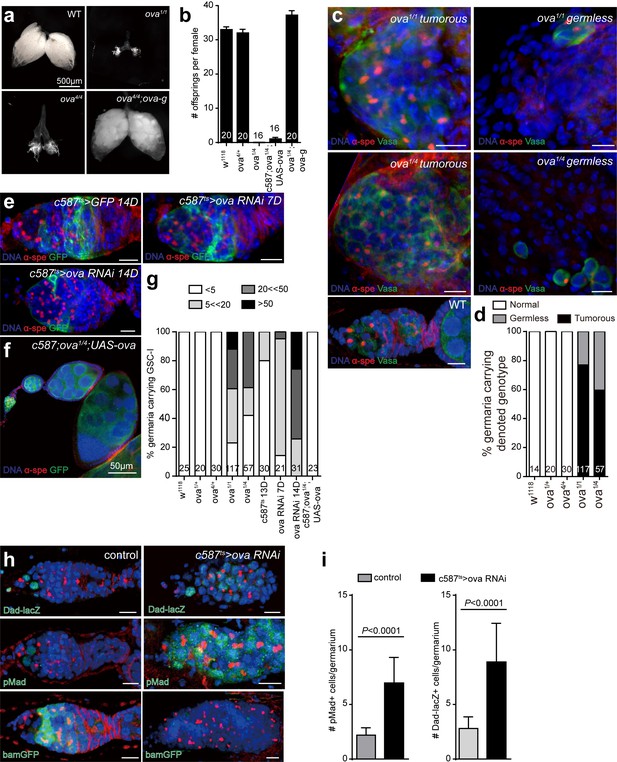
Ova is a niche factor for GSCs and ovary development in Drosophila.
(a) Phase contrast images of dissected ovaries from flies of indicated genotypes. Scale bar, 500 μm. (b) A graph shows the total offspring number of indicated females (n = 20, 20, 16, 16, 20 respectively). (c) Representative image of germaria from indicated genotypes labeled by α-Spectrin (red), Vasa (green), and DAPI (blue). ova1/1 and ova1/4 ovaries have numerous spherical-shaped spectrosome-containing cells (tumorous) or are empty of germline cells (germless), indicated by lack of germline cell marker Vasa (green). A wild-type (WT) germarium is usually 2 GSCs localized to the anterior tip. Scale bar, 10 μm. (d) A graph shows the percentage of normal, germless, and tumorous germaria of indicated genotypes (n = 14, 20, 30, 117, 57 respectively). (e) c587ts > ova RNAi germarium accumulated GSC-like cells after shift to 29°C for 7 and 14 days. Scale bars, 10 μm. (f) Escort cell-specific expression of ova rescued oogenesis and GSC differentiation defect of ova1/4 females. Red, α-Spectrin; Green, Vasa (g) Quantification of GSC-like cell number in germaria of indicated genotypes (n = 25, 20, 30, 117, 57, 30, 21, 31, 23 respectively). (h) Confocal sections of germaria stained by indicated antibodies or reporter. Scale bars, 10 µm. (i) Quantitative results of pMad and Dad-lacZ positive cell numbers from germaria of indicated genotypes. Values are mean ± SEM.; n > 20. P values by two-tailed Student t-test.
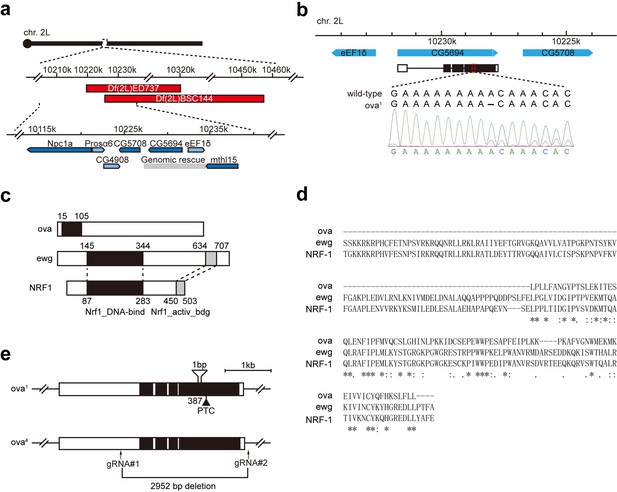
Ova is allelic to CG5694.
(a) Top, a schematic drawing of mapping results of ova1 allele using deficiency kit. Two small deficiencies, Df(2L)ED737 and Df(2L)BSC144, both failed to complement ova1 (Figure 1—figure supplement 1a). Bottom, analysis of the overlapping regions of these deficiencies implicated five candidate genes. Gray box, ova genomic rescue fragment. (b) DNA sequencing revealed a nucleotic deletion in the coding region of CG5694. (c) Schematic drawings of Ova, Drosophila Ewg-PA, and mouse NRF-1 PA proteins. Black box, Nrf1 DNA-binding domain. Grey box, Nrf1 activator-binding domain. (d) Multiple sequence alignments of Nrf1-binding domain of Ova, Ewg, and NRF-1 by Clustal Omega. (e) Schematic drawings of two CG5694/ova alleles, ova1, and ova4. ova1 is a deletion allele generated using CRISPR-Cas9.
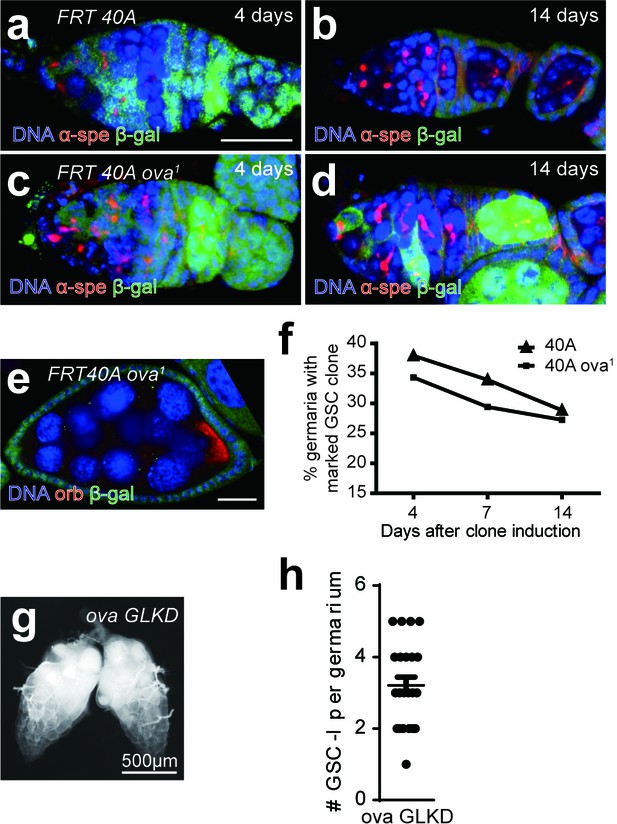
Ova is not cell- autonomously required for GSC differentiation.
(a–d) Clonal analysis of wild-type and ova mutant GSCs. Anti-α-Spectrin staining was shown in red. The control clones marked by the absence of lacZ (anti-β-galactosidase, green) at 4 days or 14 days after clone induction (ACI) (a,b). The ova1 mutant clones at 4 days or 14 days ACI (c, d). Scale bar, 10 μm. (e) A ova1 germline clone with properly specified oocyte (anti-Orb, red). Scale bar, 20 μm. (f) A time course analysis of GSC maintenance rate in wild-type and ova mutant GSC clones. (g) Phase contrast images of germline-specific knock-down ova (ova GLKD) ovary. Scale bar, 500 μm. (h) Quantification of GSC-like cell number in ova GLKD germaria. Values are mean ± SEM.; n = 24.
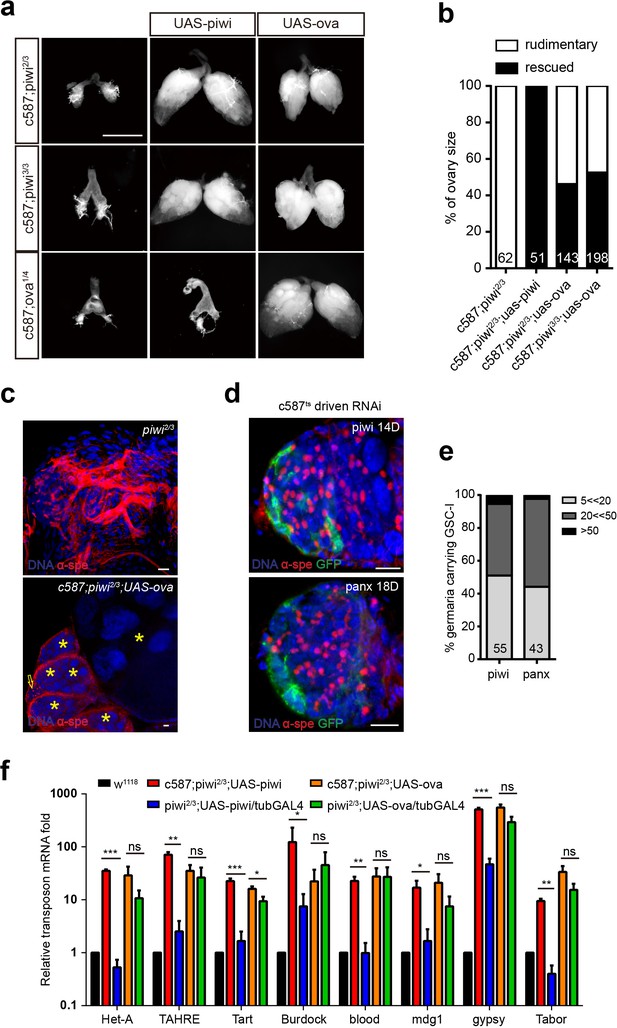
Ova acts downstream of Piwi genetically.
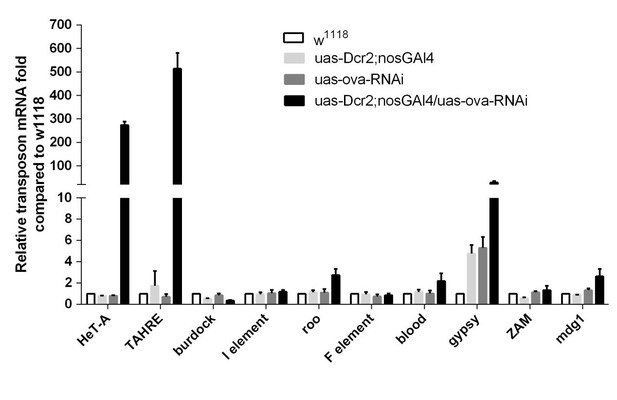
(a) Phase contrast images of dissected ovaries from flies of indicated genotypes. Scale bar, 500 µm. (b) A graph shows the percentages of germaria with rudimentary or rescued ovaries (n = 62, 51, 143, 198 respectively). (c) Confocal sections of piwi2/3 and ova-rescued ovaries. Arrow indicates the GSC-like tumor; asterisk indicates the developing germline cyst. Red, α-Spectrin. Scale bars, 10 µm. (d) Confocal sections of piwi RNAi and panx RNAi germaria. Red, α-Spectrin. Scale bars, 10 µm. (e) Quantification of GSC-like cell number in germaria of indicated genotypes. (n = 55, 43 respectively). f, qPCR result of TE levels in total ovarian RNA from indicated genotypes (normalized to actin5c). Values are means ± SEM.; n = 3. P values by two-tailed t-test (*, p<0.05; **, p<0.01; ***, p<0.001).
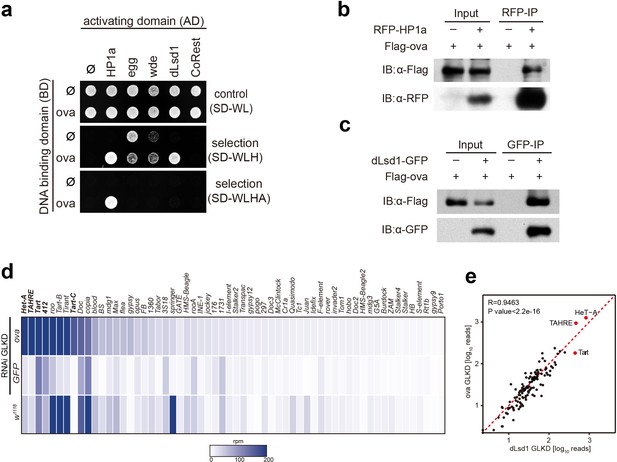
Ova interacts with the heterochromatin machinery.
(a) Y2H assay for protein interaction between Ova and proteins as indicated. (b–c) Western blots showing reciprocal co-IP between Ova and HP1a, and between Ova and dLsd1. The RFP-HP1a transgene was driven by the endogenous promoter. The dLsd1-GFP transgene was driven by a ubiquitous promoter. The Flag-ova transgene was driven by nos-GAL4. (d) Heat map displaying steady state mRNA levels as reads per million (rpm) for the top 60 detected transposons in nosGAL4 driven ova-RNAi, EGFP-RNAi, and w1118 ovaries. The average of three replicates is shown. The most upregulated transposons are highlighted in bold. (e) Correlation scatter plot of log10 transposon mRNA-seq reads between ova GLKD and dLsd1 GLKD ovaries. R = 0.9463, p<2.2×10−16 by Pearson’s correlation coefficient. The most upregulated transposons in both genotypes are highlighted in red dots.
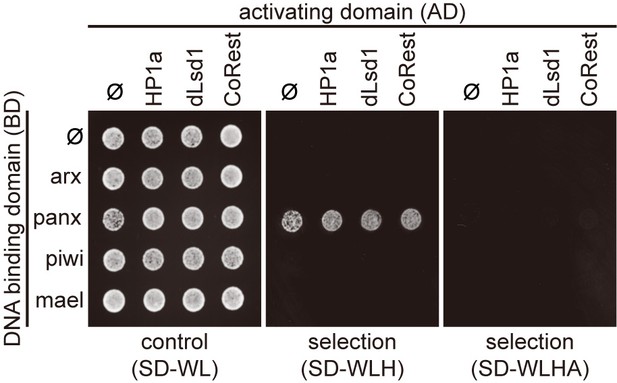
Protein interaction mapping among components of heterochromatin machinery by Y2H assay.
Y2H assay to test the interaction between proteins that associated with Piwi/piRNAs complexes and proteins in the heterochromatin machinery. SD-WL, non-selective medium; SD-WLH and SD-WLHA, selective media. Ф, empty vector.
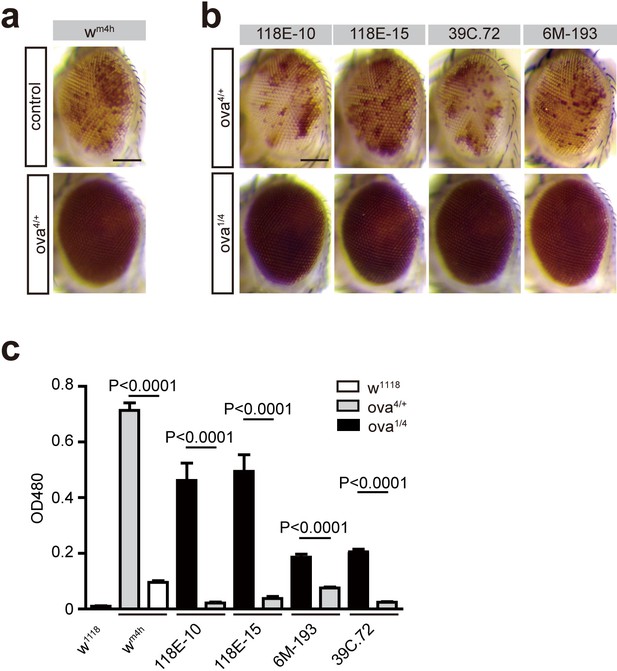
Ova acts as a suppressor of position-effect variegation (PEV).
(a) Photographs showing representative eye pigments of adult females bearing X chromosome wm4h reporter plus wild-type and ova heterozygous alleles. (b) Photographs showing representative eye pigments of adult females bearing fourth chromosome PEV reporters plus heterozygous or trans-heterozygous of ova mutant alleles. (c) Quantitative measure of eye pigment levels of indicated genotypes by a spectrophotometer. Values are mean ± SEM.; n > 5. P values by two-tailed Student’s t-test.
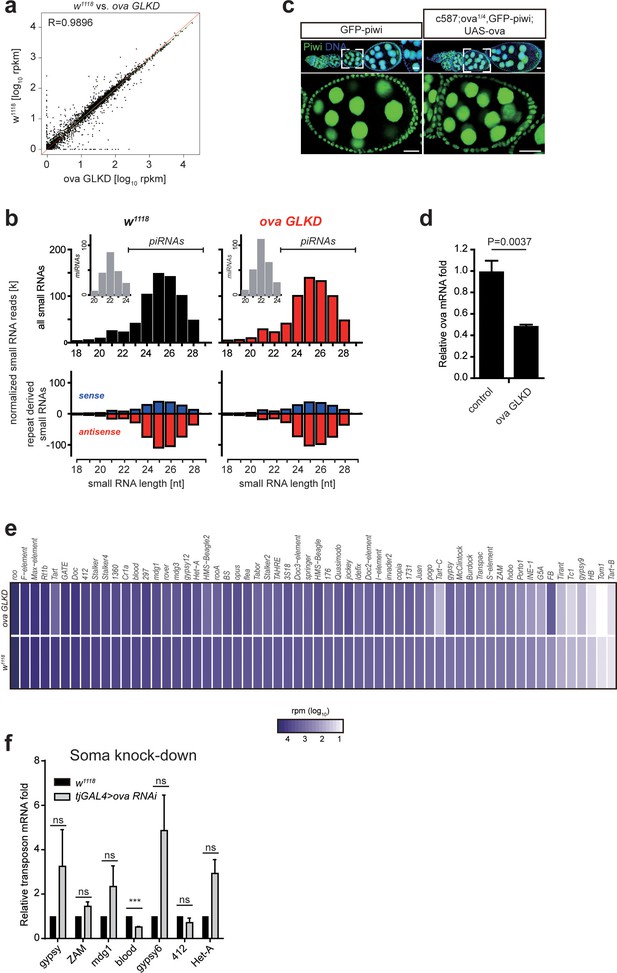
Ova is not required for piRNA biogenesis.
(a) A scatter plot shows gene expression (rpkm, reads per kilobase per million sequenced reads) comparison of protein-coding genes between w1118 and ova GLKD ovaries. R by Pearson’s correlation coefficient. (b) Top, normalized small RNA length profiles: miRNAs (gray columns, insets) and piRNAs/siRNAs (black or red columns) from ovaries with indicated genotypes. Bottom, length distribution and abundance of repeat-derived small RNAs (blue, sense; red, antisense). (c) GFP-Piwi immunostaining from ovaries of indicated genotypes. Scale bars, 10 μm. (d) qPCR result of ova mRNA levels in control and ova GLKD ovaries. Values are means ±SEM.; n = 4. P value by two-tailed Student t-test. (e) Heat map displaying repeat small RNA levels as reads per million (rpm) for individual transposon in ova GLKD and w1118 ovaries. (f) qPCR result of TE mRNA levels in w1118 and tjGAL4 >ova RNAi (shift to 29°C for 7 days) ovaries. ***, p<0.001 by two-tailed Student’s t-test. Values are means ± SEM.; n = 3.
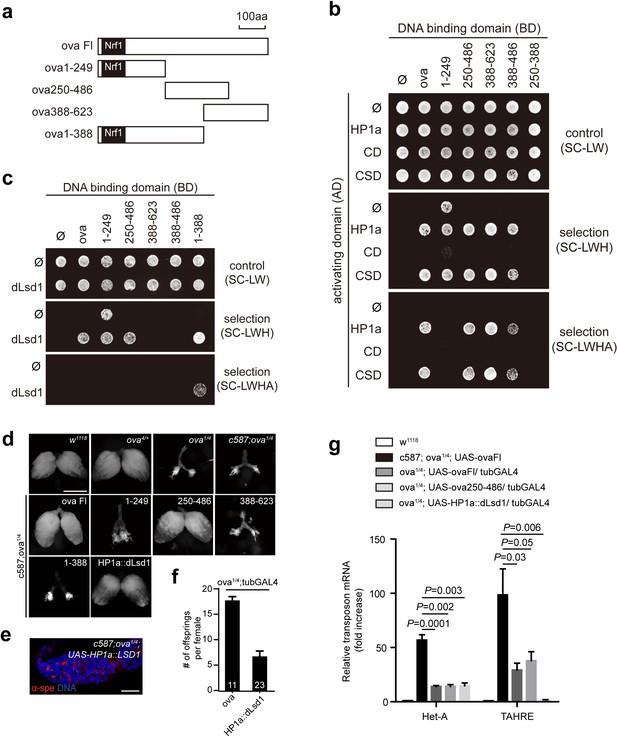
Ova acts as a protein adaptor to link dLsd1 with HP1a.
(a) Schematic drawings of full length and truncated forms of Ova. (b) Mapping the reciprocal-binding regions between HP1a and Ova by Y2H assay. (c) Mapping the reciprocal-binding regions between HP1a and dLsd1 by Y2H assay. (d) Ovaries from flies of indicated genotypes. Escort cell-specific expression of ova full length, ova250-486 or HP1a::dLsd1 rescued ova1/4 ovary defect. Scale bar, 500 µm. (e) A representative image of ova1/4 germarium rescued by escort cell-specific expression of HP1a::dLsd1. Red, α-spectrin; Blue, DAPI. Scale bar, 10 µm. (f) A graph shows the total offspring number of indicated females (n = 11, 23 respectively). (g) A graph shows fold changes of TEs in total ovarian RNA from indicated genotypes (normalized to actin5c). Values are means ± SEM.; n > 4. P values by two-tailed t-test.
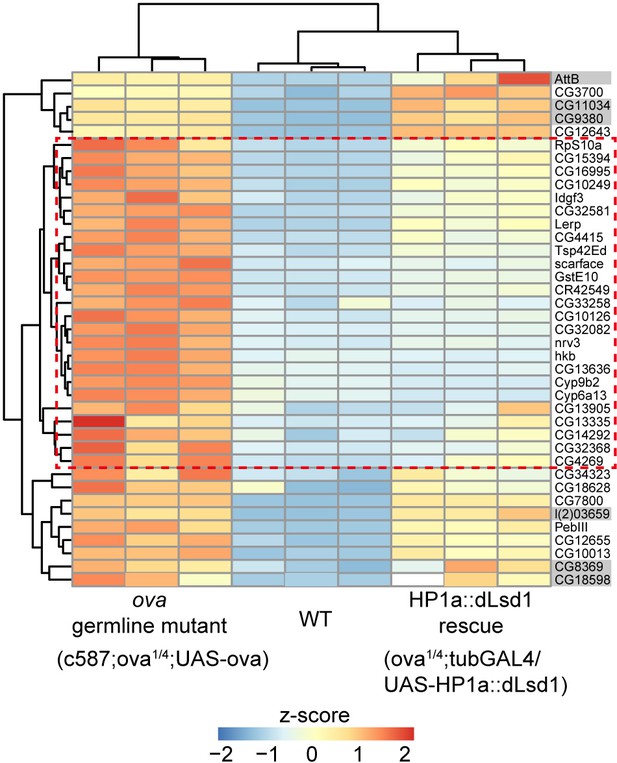
HP1a::dLsd1 expression rescues ova loss induced derepression of protein-coding genes.
Heat map showing relative z-score of mRNA-seq from indicated genotype ovaries. The top 39 upregulated genes in ova germline mutant ovaries are shown and the genes, which are rescued to wild-type levels by HP1a::dLsd1, are highlighted in red dashed box. Grey shadow, not significant.
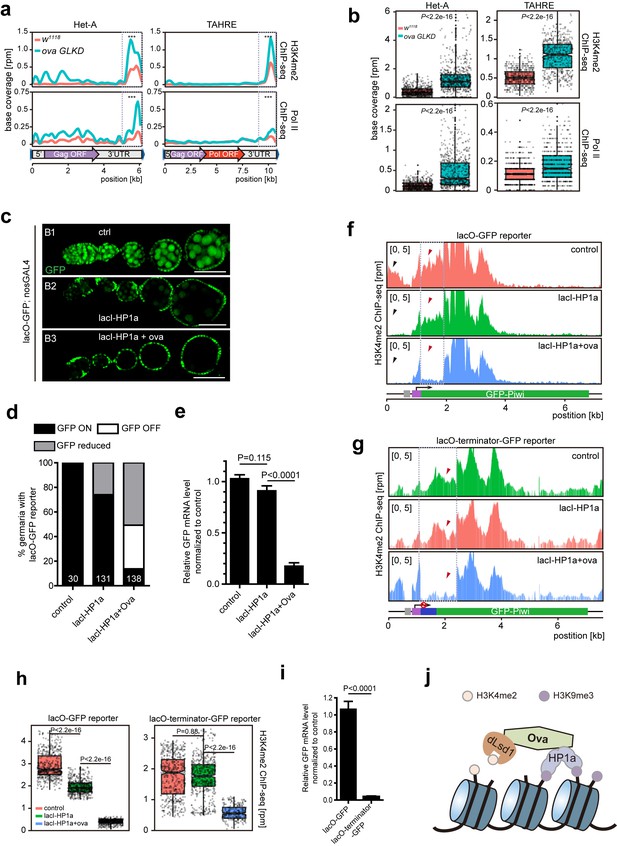
Ova regulates HP1a-induced local H3K4 demethylation.
(a) Graphs showing H3K4me2 and Pol II ChIP-seq profiles mapped to indicated transposon loci in control versus ova GLKD ovaries. Dashed boxes, enhancer regions of transposons. RPM, reads per million. Bin, 100 bp. (b) Quantitative comparison of H3K4me2 and Pol II densities in the indicated enhancer regions in 5a (dashed boxes). P values by two-tailed Student’s t-test. (c), Ovarioles with indicated genotypes expressing ubiquitous lacO-GFP reporter in the germline cells. GFP was visualized by antibody staining. Scale bar, 50 µm. (d) A graph showing the percentage of ovarioles of indicated genotypes with normal, reduced, or abolished GFP signals. (e) Quantitative RT-PCR results of GFP mRNA from ovaries of indicated genotypes. Values are means ±SEM.; n > 4. P values by two-tailed Student’s t-test. (f) Graphs showing normalized H3K4me2 density mapped to lacO-GFP reporter region from ovaries of indicated genotypes. Grey box, lacO-binding sites; Purple box, nanos promoter. (g) Graphs showing normalized H3K4me2 density mapped to lacO-terminator-GFP reporter region from ovaries of indicated genotypes. Blue box, VASA terminator. (h) Quantitative comparison of H3K4me2 density in regions indicated by the dashed boxes in f or g. P values by two-tailed Student’s t-test. (i) Quantitative RT-PCR results of GFP mRNA from lacO-GFP and lacO-terminator-GFP reporter ovaries. Values are means ± SEM.; n = 4. P value by Student t-test. (j) A schematic model for Ova function: Ova functions as a protein adaptor to link HP1a with dLsd1 for local H3K4 demethylation during HP1a-induced transcriptional gene silencing.
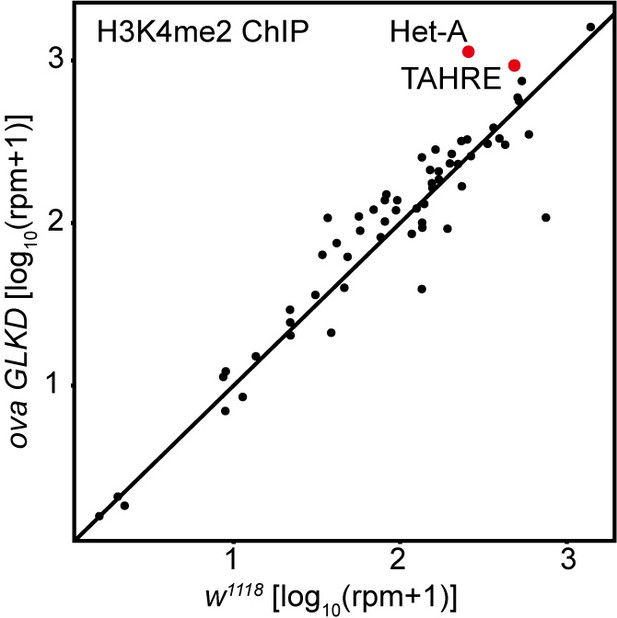
Comparison of H3K4me2 density on all transposons in w1119 and ova GLKD ovaries.
Correlation scatter plot of log10 H3K4me2 ChIP-seq reads between w1118 and ova GLKD ovaries. The Het-A and TAHRE transposons are highlighted in red dots.
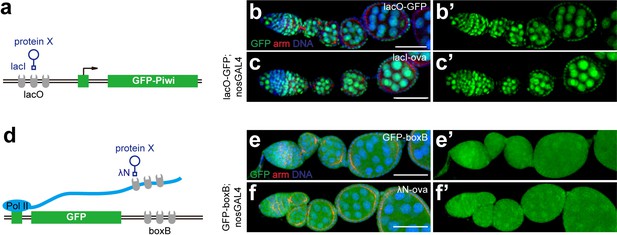
Tethering Ova to DNA or RNA is unable to induce co-transcriptional gene silencing.
(a), A cartoon to describe the DNA tethering assay using the lacI and lacO binary system. (b, b’, c, c’) Confocal images of GFP expression in ovarioles of indicated genotypes. Green, anti-GFP staining. Red, anti-Armadillo. (d), A cartoon to describe the RNA tethering assay using λN and boxB binary system. (e, e’, f, f’) Confocal images of GFP expression in ovarioles of indicated genotypes. Scale bars, 50 μm.

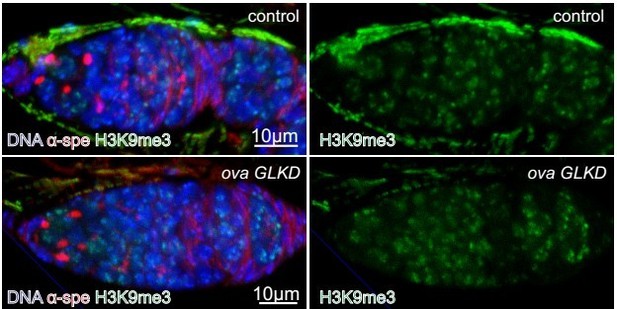
dLsd1 and HP1a expression and subcellular localization in control and ova GLKD germaria
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Drosophila melanogaster) | ova[1] | This paper | See Materials and methods | |
| Genetic reagent (Drosophila melanogaster) | ova[4] | This paper | See Materials and methods | |
| Genetic reagent (Drosophila melanogaster) | c587-GAL4 | (Song et al., 2004) (DOI: 10.1242/dev.01026) | RRID:BDSC_67747 | |
| Genetic reagent (Drosophila melanogaster) | Dad-lacZ | (Tsuneizumi et al., 1997) (DOI: 10.1038/39362) | RRID:DGGR_118114 | |
| Genetic reagent (Drosophila melanogaster) | bam-GFP | (Chen and McKearin, 2003) | RRID:DGGR_118177 | |
| Genetic reagent (Drosophila melanogaster) | piwi[2] | (Lin and Spradling, 1997) | RRID:BDSC_43319 | |
| Genetic reagent (Drosophila melanogaster) | piwi[3] | (Lin and Spradling, 1997) | RRID:BDSC_12225 | |
| Genetic reagent (Drosophila melanogaster) | GFP-piwi | Katalin Toth (California Institute of Tchnology) | ||
| Genetic reagent (Drosophila melanogaster) | 118E-10 | Lori Wallrath (University of Iowa) | ||
| Genetic reagent (Drosophila melanogaster) | 118E-15 | Lori Wallrath (University of Iowa) | ||
| Genetic reagent (Drosophila melanogaster) | 6 M-193 | Lori Wallrath (University of Iowa) | ||
| Genetic reagent (Drosophila melanogaster) | 39C.72 | Lori Wallrath (University of Iowa) | ||
| Genetic reagent (Drosophila melanogaster) | dLsd1-GFP | Yu Yang (Institute of Biophysics, Chinese Academy of Science) | ||
| Genetic reagent (Drosophila melanogaster) | EGFP-RNAi | Bloomington Drosophila Stock Center | (#41553) | |
| Genetic reagent (Drosophila melanogaster) | RFP-HP1a | Bloomington Drosophila Stock Center | (#30562) | |
| Genetic reagent (Drosophila melanogaster) | UAS-Dcr2; nos-GAL4 | Bloomington Drosophila Stock Center | (#25751) | |
| Genetic reagent (Drosophila melanogaster) | tub-GAL4 | Bloomington Drosophila Stock Center | (#5138) | |
| Genetic reagent (Drosophila melanogaster) | tub-GAL80ts | Bloomington Drosophila Stock Center | (#7016, #7018) | |
| Genetic reagent (Drosophila melanogaster) | Df(2L)BSC144 | Bloomington Drosophila Stock Center | (#9504) | |
| Genetic reagent (Drosophila melanogaster) | attP2 | Bloomington Drosophila Stock Center | (#25710) | |
| Genetic reagent (Drosophila melanogaster) | In(1)wm4h | Kyoto Stock Center | (#101652) | |
| Genetic reagent (Drosophila melanogaster) | Df(2L)ED737 | Kyoto Stock Center | (#150520) | |
| Genetic reagent (Drosophila melanogaster) | ova-RNAi | Vienna Drosophila Research Center | (#102156) | |
| Genetic reagent (Drosophila melanogaster) | piwi-RNAi | Vienna Drosophila Research Center | (#101658) | |
| Genetic reagent (Drosophila melanogaster) | panx-RNAi | Vienna Drosophila Research Center | (#102702) | |
| Genetic reagent (Drosophila melanogaster) | EGFP-5xBoxB | Vienna Drosophila Research Center | (#313408) | |
| Genetic reagent (Drosophila melanogaster) | lacO-GFP-Piwi | Vienna Drosophila Research Center | (#313394) | |
| Genetic reagent (Drosophila melanogaster) | lacI-HP1a; lacO- GFP-Piwi | Vienna Drosophila Research Center | (#313409) | |
| Genetic reagent (Drosophila melanogaster) | nos-Cas9 | Jianquan Ni (Tsinghua University) | ||
| Genetic reagent (Drosophila melanogaster) | pCasper4-ova-g | This paper | See Materials and methods | |
| Genetic reagent (Drosophila melanogaster) | GFP-ova | This paper | See Materials and methods | |
| Genetic reagent (Drosophila melanogaster) | UASP-ova | This paper | See Materials and methods | |
| Genetic reagent (Drosophila melanogaster) | UASP-piwi | This paper | See Materials and methods | |
| Genetic reagent (Drosophila melanogaster) | UASP-ova1-249 | This paper | See Materials and methods | |
| Genetic reagent (Drosophila melanogaster) | UASP-ova250-486 | This paper | See Materials and methods | |
| Genetic reagent (Drosophila melanogaster) | UASP-ova388-623 | This paper | See Materials and methods | |
| Genetic reagent (Drosophila melanogaster) | UASP-ova1-388 | This paper | See Materials and methods | |
| Genetic reagent (Drosophila melanogaster) | UASP-HP1a::dLsd1 | This paper | See Materials and methods | |
| Genetic reagent (Drosophila melanogaster) | UASP-λN-ova | This paper | See Materials and methods | |
| Genetic reagent (Drosophila melanogaster) | UASP-lacI-ova | This paper | See Materials and methods | |
| Genetic reagent (Drosophila melanogaster) | lacO-terminator-GFP-Piwi | This paper | See Materials and methods | |
| Recombinant DNA reagent | UASP-λN | Julius Brennecke (Institute of Molecular Biotechnology) | ||
| Recombinant DNA reagent | UASP-lacI | |||
| Recombinant DNA reagent | lacO-GFP-Piwi | |||
| Recombinant DNA reagent | pGBKT7 | Clontech (Cat#630443) | ||
| Recombinant DNA reagent | pGAD | Clontech (Cat#630442) | ||
| Antibody | rabbit polyclonal anti-pMad | Ed Laufer (Columbia Universtity Medical Center) | RRID:AB_2617125 | IHC(1:1000) |
| Antibody | rabbit polyclonal anti-β-galactosidase | MP Biologicals (Cat#0855976) | RRID:AB_2687418 | IHC(1:3000) |
| Antibody | mouse monoclonal anti-α-Spectrin | Developmental Studies Hybridoma Bank | IHC(1:50) | |
| Antibody | mouse monoclonal anti-Tubulin | Developmental Studies Hybridoma Bank | RRID:AB_1157911 | WB(1:2000) |
| Antibody | rabbit polyclonal anti-mCherry | BioVision (cat#5993) | RRID:AB_1975001 | WB (1:2000) |
| Antibody | rabbit polyclonal anti-GFP | Life (cat#A11122) | RRID:AB_221569 | IHC(1:1000) WB(1:10000) |
| Antibody | polyclonal anti-rabbit IgG-HRP | ZSJQ-BIO (cat#ZB2301) | WB(1:10000) | |
| Antibody | rabbit polyclonal anti-H3K4me2 | Abcam (cat#ab7766) | RRID:AB_732924 | |
| Antibody | mouse monoclonal anti-RNA polymerase II | Abcam (cat#ab817) | RRID:AB_306327 | |
| Antibody | mouse monoclonal anti-Flag | Sigma (cat#F1804) | RRID:AB_439685 | IHC(1:300) WB(1:6000) |
| Chemical compound, drug | 4’,6’-diamidino-2- phenylindole | Sigma (cat#10236276001) | ||
| Commercial assay or kit | anti-Flag resin | Sigma (cat#A2220) | RRID:AB_10063035 | |
| Commercial assay or kit | GFP-Trap agarose | Chromoteck (cat#gta-10) | ||
| Commercial assay or kit | RFP-Trap agarose | Chromoteck (cat#rta-10) | ||
| Commercial assay or kit | Qiagen Plasmid Midi Kit | Qiagen (#12145) | ||
| Commercial assay or kit | Immobilon Western Chemiluminescent HRP Substrate Kit | Millipore (cat#WBKLS0500) | ||
| Commercial assay or kit | HiScript II Q RT SuperMix | Vazyme Biotech (cat#R223-01) | ||
| Commercial assay or kit | ChamQ SYBR qPCR master Mix | Vazyme Biotech (cat#Q331) | ||
| Commercial assay or kit | Oligo d(T)25 Magnetic beads | NEB (cat#S1419S) | ||
| Commercial assay or kit | NEBNext Ultra IIDNA Library Prep Kits for Illumina | NEB (cat# E7645S) | ||
| Commercial assay or kit | VAHTS Small RNA Library Prep Kit for Illumina | Vazyme Biotech (cat#NR801) | ||
| Commercial assay or kit | VAHTS Universal DNA Library Prep Kit | Vazyme Biotech (cat#ND607) | ||
| Commercial assay or kit | TruePrep Index Kit | Vazyme Biotech (cat#TD202) | ||
| Software, algorithm | GraphPad Prism | GraphPad Prism (https://graphpad.com) | RRID:SCR_002798 | |
| Sequenced-based reagent | RT-qPCR primers | This paper | See Supplementary file 1, Table 2 |
Additional files
-
Supplementary file 1
Table 1: Viability test of ova mutants. Table 2: Primers used in this study.
- https://doi.org/10.7554/eLife.40806.016
-
Transparent reporting form
- https://doi.org/10.7554/eLife.40806.017