Single-nucleus transcriptomics reveal the differentiation trajectories of periosteal skeletal/stem progenitor cells in bone regeneration
Figures

Heterogeneity of the periosteum at steady state.
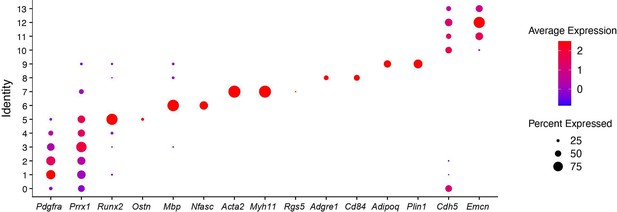
(A) Experimental design. Nuclei were extracted from the periosteum of uninjured tibia and processed for single-nucleus RNAseq. (B) Sorting strategy of nuclei stained with Sytox-7AAD for snRNAseq. Sorted nuclei are delimited by a red box. (C) UMAP of color-coded clustering of the uninjured periosteum dataset. Eight populations are identified and delimited by black dashed lines. (D) Violin plots of key marker genes of the different cell populations.

Identification of periosteal skeletal stem/progenitor cells in the intact periosteum.
(A) UMAP of color-coded clustering of the subset of SSPCs/fibroblasts. (B) Feature plots of Prrx1 and Pdgfra in the subset of SSPCs/fibroblasts. (C) Feature plots of key marker genes of the different cell populations. (D) Dot plot of the stemness markers Pi16, Ly6a (SCA1), Cd34, and Dpp4. (E) Violin and feature plots of CytoTrace scoring in the subset of SSPCs/fibroblasts, showing that SCA1 expressing SSPCs (cluster 0) are the less differentiated cells in the dataset. (F) Experimental design: GFP+ SCA1+ and GFP+ SCA1- were isolated from uninjured tibia of Prrx1Cre; R26mTmG mice and used for in vitro CFU assays or grafted at the fracture site of wild-type mice. (G) In vitro CFU assay of murine periosteal Prrx1-GFP+ SCA1+ and Prrx1-GFP+ SCA1- cells (n = 5 biological replicates from 2 distinct experiments). (H) High magnification of SOX9 immunofluorescence of callus section 14 days post-fracture showing that GFP+SCA1+ cells contribute to the callus (white arrowheads) while GFP+ SCA1- cells are not contributing (n = 3 per group).

Expression of known skeletal stem/progenitor cell (SSPC) markers in the periosteum at steady state.
Feature plots of known markers of SSPCs in the SSPC/fibroblast subset of the periosteum at steady state.

Periosteal response to fracture at single-nucleus resolution.
(A) Experimental design. Nuclei were extracted from the periosteum of uninjured tibia and from the injured periosteum and hematoma/callus at days 3, 5, and 7 post-tibial fracture of wild-type mice and processed for single-nucleus RNAseq. (B) UMAP of color-coded clustering of the integration of uninjured, day 3, 5, and 7 datasets. Eleven populations are identified and delimited by black dashed lines. (C) Violin plots of key marker genes of the different cell populations. (D) UMAP of the combined dataset separated by time point. (E) Percentage of cells in skeletal stem/progenitor cell (SSPC), injury-induced fibrogenic cell, osteoblast, chondrocyte, and immune cell clusters in uninjured, day 3, 5, and 7 datasets.
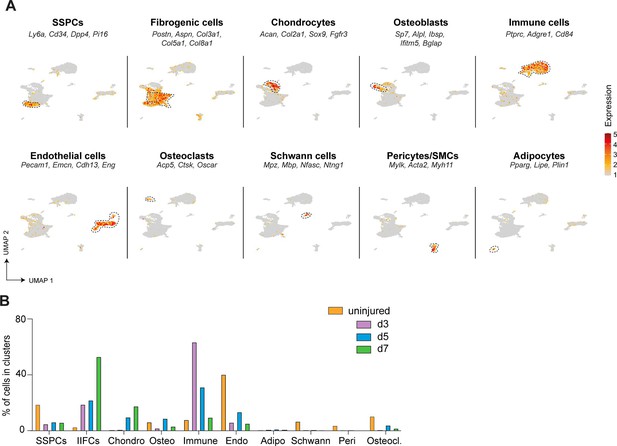
Heterogeneity and dynamics of the cell populations in the fracture environment.
(A) Feature plots of the lineage score of the different cell populations in the combined fracture datasets. (B) Percentage of cells in each cell population per time point.
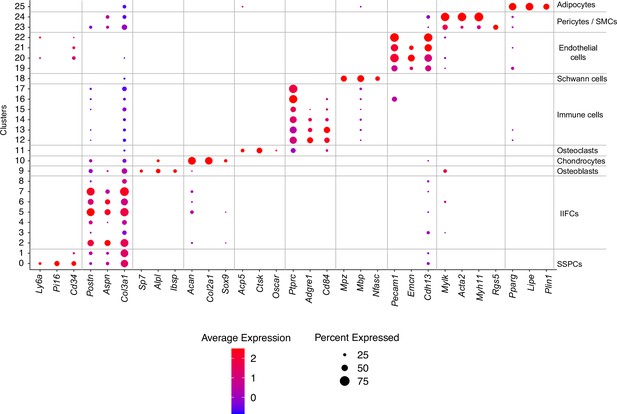
Dot plot of marker genes of the populations from the combined fracture dataset.

Cellular organization of the fracture callus.
(A) Picrosirius staining of the uninjured periosteum. (B) Immunofluorescence and RNAscope on adjacent sections show the presence of SSPCs (Pi16-expressing cells) in the fibrous layer (fl), Postn-expressing cells in the cambium layer (cl), OSX+ osteoblasts, immune cells (CD45+), and endothelial cells (PECAM1+) in the periosteum (n = 3 per group). (C) Safranin’O staining of longitudinal callus sections at day 3 post-tibial fracture. (D) Immunofluorescence and RNAscope on adjacent sections show absence of skeletal stem/progenitor cells (SSPCs) (Pi16+), and presence of IIFCs (Postn+) and immune cells (CD45+) in the activated periosteum and hematoma at day 3 post-fracture. Chondrocytes (SOX9+, white arrowhead), osteoblasts (OSX+,white arrowhead), immune cells (CD45+), and endothelial cells (PECAM1+) are detected in the activated periosteum (n = 3 per group). (E) Safranin’O staining of longitudinal callus sections at day 5 post-tibial fracture. (F) Immunofluorescence and RNAscope on adjacent sections show injury-induced fibrogenic cells (IIFCs) (Postn+), chondrocytes (SOX9+, white arrowhead), osteoblasts (OSX+, white arrowhead), immune cells (CD45+), and endothelial cells (PECAM1+) in the fibrosis, chondrocytes (SOX9+) in the cartilage and osteoblasts (OSX+), immune cells (CD45+,white arrowhead), and endothelial cells (PECAM1+) in the new bone (n = 3 per group). (G) Safranin’O staining of longitudinal callus sections at day 7 post-tibial fracture. (H) Immunofluorescence and RNAscope on adjacent sections show IIFCs (Postn+), chondrocytes (SOX9+, white arrowhead), osteoblasts (OSX+, white arrowhead), immune cells (CD45+), and endothelial cells (PECAM1+) in the fibrosis, chondrocytes (SOX9+) in the cartilage and osteoblasts (OSX+), immune cells (CD45+, white arrowhead), and endothelial cells (PECAM1+) in the new bone (n = 3 per group). Scale bars: (A–B–E) 1 mm, (B–D–F) 100 µm.
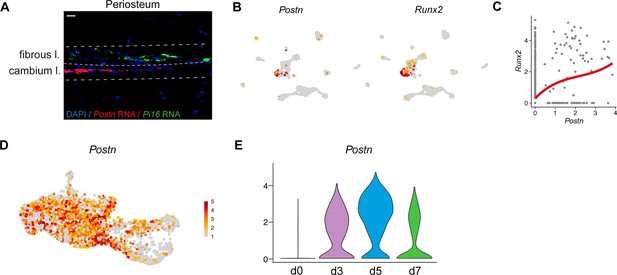
Periosteal skeletal stem/progenitor cells do not express Postn.
(A) RNAscope experiment showing the presence of Postn expressing cells in the inner cambium layer (cambium l.) of the periosteum and Pi16-expressing cells in the fibrous layer (fibrous l.). (B) Feature plots of Postn and Runx2 expression in the uninjured periosteum. (C) Scatter plot of Runx2 and Postn expression in the uninjured periosteum dataset showing that Postn is mostly expressed by cells expressing Runx2. (D) Feature plot of Postn expression in the subset of SSPCs, injury-induced fibrogenic cells (IIFCs), osteogenic and chondrogenic cells from Figure 5B. (E) Violin plot of Postn expression per time point.
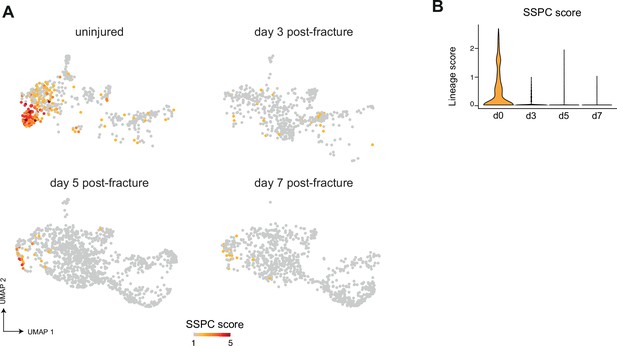
Absence of skeletal stem/progenitor cells in the injured periosteum.
(A) Feature plot of SSPC lineage score in the subset of SSPCs, injury-induced fibrogenic cells (IIFCs), osteoblasts, and chondrocytes separated by time point from Figure 5B. (B) Violin plot of SSPC lineage score by time point.

Periosteal skeletal stem/progenitor cells activate through a common fibrogenic state prior to undergoing osteogenesis or chondrogenesis.
(A) SSPCs, injury-induced fibrogenic cells (IIFCs), chondrocytes, and osteoblasts from integrated uninjured, day 3, 5, and 7 post-fracture samples were extracted for a subset analysis. (B) UMAP of color-coded clustering (left), color-coded sampling (middle), and monocle pseudotime trajectory (right) of the subset dataset. The four populations are delimited by black dashed lines. (C) (Top) Feature plots of the stem/progenitor, fibrogenic, chondrogenic, and osteogenic lineage scores. (Middle) Scatter plot of the lineage scores along pseudotime. (Bottom) Violin plot of the lineage scores per time point. (D) Distribution of the cells along the pseudotime per time point. (E) Schematic representation of the differentiation trajectories of pSSPCs after fracture.
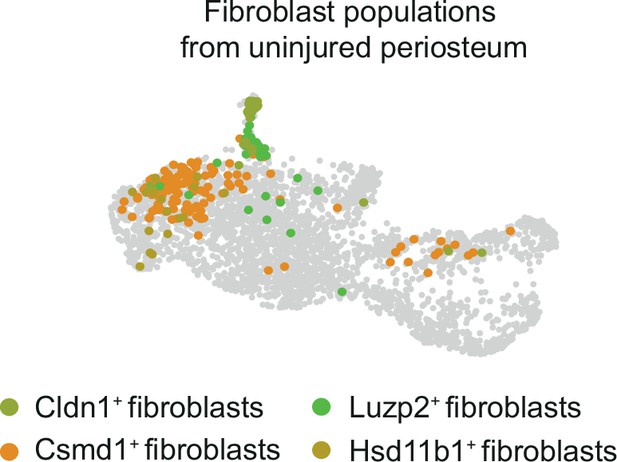
UMAP highlighting the distribution of periosteal fibroblasts in the combined fracture dataset.

In vivo validation of periosteal skeletal stem/progenitor cell differentiation trajectories.
(A) (Top) Representative Safranin’O staining on longitudinal sections of the hematoma/callus at day 5 post-fracture. The callus is composed of fibrosis, cartilage (red dashed line), and bone (green dashed line). (Middle, box 1) Immunofluorescence on adjacent section shows decreased expression of POSTN (green) and increased expression of SOX9 (red) in the fibrosis-to-cartilage transition zone. (Bottom, box 2) Immunofluorescence on adjacent section shows decreased expression of POSTN (green) and increased expression of OSX (red) in the fibrosis-to-bone transition zone (n = 3 per group). (B) Experimental design: GFP+ SCA1+ SSPCs were isolated from uninjured tibia of Prrx1Cre; R26mTmG mice and grafted at the fracture site of wild-type mice. Safranin’O staining of callus sections at day 5 post-fracture and high magnification of POSTN immunofluorescence of adjacent section showing that grafted GFP+ SSPCs contribute to the callus and differentiate into POSTN+ IIFCS (white arrowheads) (n = 4 per group). (C) Experimental design: GFP+ IIFCs from periosteum and hematoma at day 3 post-fracture tibia were isolated from Prrx1Cre; R26mTmG mice and grafted at the fracture site of wild-type mice. Safranin’O of callus sections at day 14 post-fracture and high magnification of OSX and SOX9 immunofluorescence of adjacent sections showing that grafted GFP+ injury-induced fibrogenic cells (IIFCs) contribute to the callus and differentiate into OSX+ osteoblasts (box 3, white arrowheads) and SOX9+ chondrocytes (box 4, white arrowheads) (n = 4 per group). Scale bars: low magnification: (A) 500 µm; (B, C) 1 mm. High magnification: 100 µm.
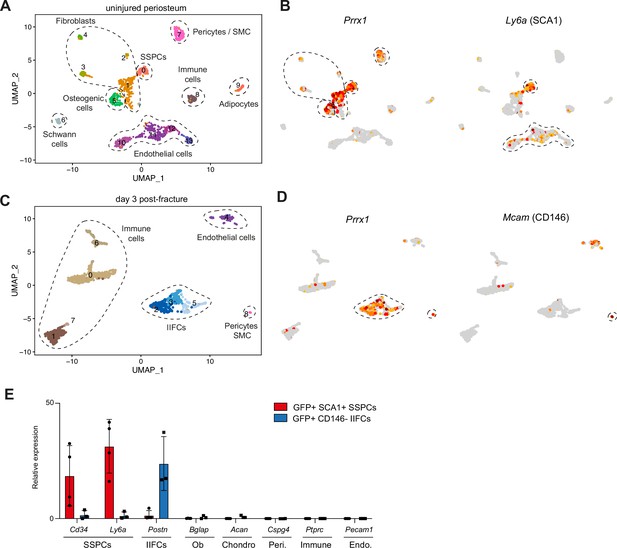
Validation of skeletal stem/progenitor cell (SSPC) and injury-induced fibrogenic cell (IIFC) sorting strategies.
(A) UMAP of the clustering of the uninjured periosteum. (B) Feature plots of Prrx1 and Ly6a expression in the uninjured dataset. (C) UMAP of the clustering of the day 3 post-fracture periosteum and hematoma. (D) Feature plots of Prrx1 and Mcam (CD146) expression in the day 3 post-fracture periosteum and hematoma dataset. (E) Relative expression of cell population markers by GFP+ SCA1+ SSPCs from uninjured periosteum (n = 4) and GFP+ CD146- IIFCs from day 3 post-fracture periosteum and hematoma (n = 3).

Characterization of injury-induced fibrogenic cells.
(A) Gene Ontology analyses of upregulated genes in injury-induced fibrogenic cells (IIFCs) (clusters 2–6 of UMAP clustering from Figure 5). (B) Dot plot of extracellular matrix (ECM) genes in UMAP clustering from Figure 5. (C) Feature plot per cluster and scatter plot along pseudotime of the mean expression of ECM genes. (D) Gene regulatory network (GRN)-based tSNE clustering of the subset of skeletal stem/progenitor cells (SSPCs), IIFCs, chondrocytes, and osteoblasts. (E) Activation of Mta3, Six1, Sox9, and Sp7 regulons in SSPCs, IIFCs, chondrocytes, and osteoblasts. Blue dots mark cells with active regulon. (F) Number of regulons activated per cell in the SSPC, IIFC, osteoblast (Ob), and chondrocyte (Ch) populations. Statistical differences were calculated using one-way ANOVA. ***p-value<0.001. (G) Heatmap of activated regulons in SSPC, IIFC, osteoblast (osteob), and chondrocyte (chondro) populations. (H) Scatter plot of the activity of the combined fibrogenic regulons along monocle pseudotime from Figure 5. (I) Reactome pathway analyses of the fibrogenic regulons shows that the three most significant terms are related to Notch signaling (blue). (J) Feature plot in Seurat clustering and scatter plot along monocle pseudotime of the Notch signaling score.
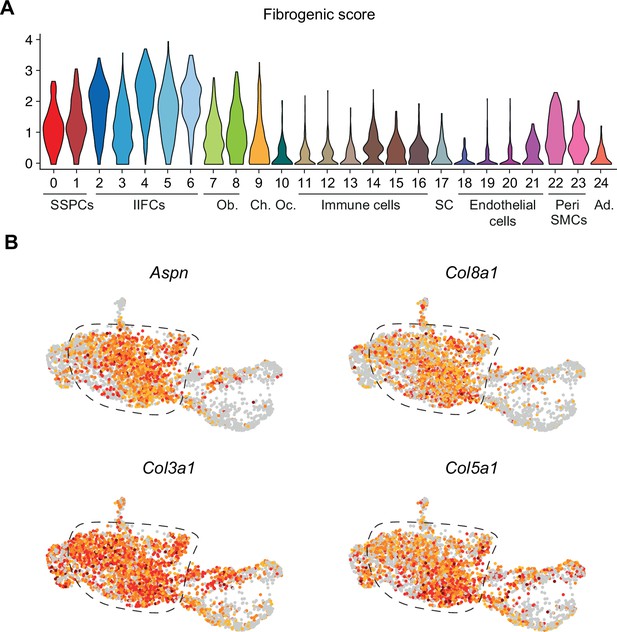
Injury-induced fibrogenic cells (IIFCs) are expressing extracellular matrix (ECM)-related genes.
(A) Violin plot of the fibrogenic score in the integrated dataset. (B) Feature plots of Aspn, Col3a1, Col5a1, and Col8a1 in the subset of skeletal stem/progenitor cells (SSPCs), IIFCs, osteoblasts, and chondrocytes.
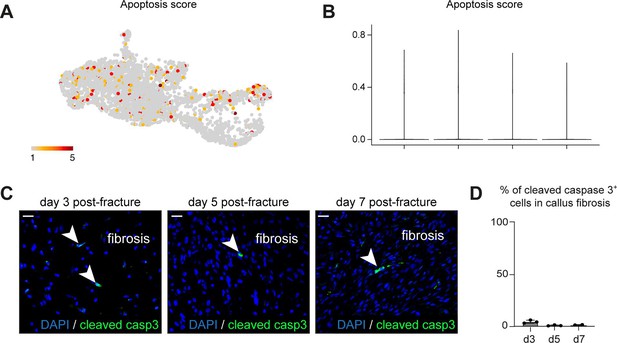
Injury-induced fibrogenic cells (IIFCs) do not undergo apoptosis.
(A) Feature plot of the apoptosis score in the subset of skeletal stem/progenitor cells (SSPCs), IIFCs, osteoblast, and chondrocytes. (B) Violin plot of the apoptosis score separated by time points. (C) Immunofluorescence of cleaved caspase 3 in callus fibrosis at days 3, 5, and 7 post-fracture. (D) Percentage of cleaved caspase 3 positive cells in the fibrosis callus fibrosis at day 3 (n = 3), day 5 (n = 3), and day 7 post-fracture (n = 2).

Activity of lineage specific regulons (Mta3, Six1, Sox9 and Sp7) in the UMAP Seurat clustering of SSPCs, IIFCs, chondrocytes and osteoblasts.

Gene regulatory network analyses identify gene cores driving fibrogenic to chondrogenic and osteogenic transitions.
(A) Activation of Maf, Arntl, and Nfatc2 regulons in skeletal stem/progenitor cells (SSPCs), injury-induced fibrogenic cells (IIFCs), chondrocytes, and osteoblasts. (B) STRING interaction network of the chondro-core 1 and 2 transcription factors (blue and orange, respectively). (C) Feature plot of chondro-core 1 (top) and chondro-core 2 (bottom) activities in SSPCs, IIFCs, chondrocytes, and osteoblasts in Seurat UMAP from Figure 5. (D) Scatter plot of chondro-core 1 (top) and chondro-core 2 (bottom) activities along monocle pseudotime and Acan expression. (E) Activation of Tcf7, Bclb11b, and Tbx2 regulons in SSPCs, IIFCs, chondrocytes, and osteoblasts. (F) STRING interaction network of the osteo-core transcription factors (green) and their related genes shows that most of osteo-core related genes are involved in Wnt pathway (purple). (G) Feature plot of the osteo-core activity in SSPCs, IIFCs, chondrocytes, and osteoblasts in Seurat UMAP from Figure 5. (H) Scatter plot of osteo-core activity along monocle pseudotime and Ibsp expression.
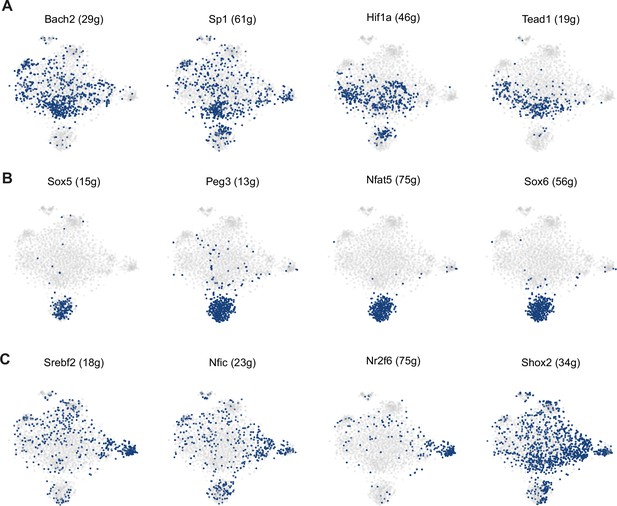
Regulon activity in the subset of skeletal stem/progenitor cells (SSPCs), injury-induced fibrogenic cells (IIFCs), osteoblasts, and chondrocytes.
(A) Activity of chondro-core 1 regulons in SCENIC tSNE clustering of SSPCs, IIFCs, chondrocytes, and osteoblasts. (B) Activity of chondro-core 2 regulons in SCENIC tSNE clustering of SSPCs, IIFCs, chondrocytes, and osteoblasts. (C) Activity of osteo-core regulons in SCENIC tSNE clustering of SSPCs, IIFCs, chondrocytes, and osteoblasts.

Injury-induced fibrogenic cells are the main source of paracrine factors after fracture.
(A) Outgoing interaction strengths of the different cell populations of the fracture environment determined using CellChat package. (B) Comparison of incoming and outgoing interaction strengths across SSPC, IIFC, chondrogenic, and osteogenic populations. (C) Outgoing and incoming signaling from and to SSPCs, IIFCs, chondrocytes, and osteoblasts. (D) Cell–cell interactions identified between SSPCs, IIFCs, chondrocytes, and osteoblasts. (E) Violin plots of the score of BMP, TGFβ, PDGF, POSTN, PTN, and ANGPTL signaling per time point. (F) Scatter plot along pseudotime and violin plot per time point of the mean expression of the ligand and receptors involved in signaling from IIFCs. (G) Circle plot of the interactions between SSPCs, IIFCs, chondrocytes, and osteoblasts, showing that most signals received by SSPCs are coming for IIFCs. Ob: osteoblasts; Oc: osteoclasts; Ch: chondrocytes; SC: Schwann cells; Ad: adipocytes.
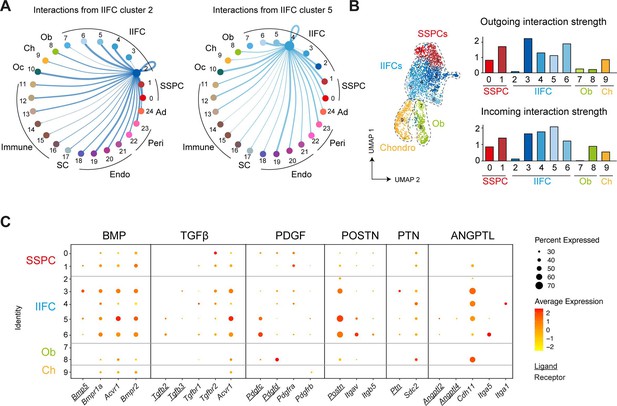
Paracrine interactions from injury-induced fibrogenic cells (IIFCs).
(A) Circle plots showing the interaction strengths between IIFCs in clusters 2 and 5 with the other cell populations. (B) (Left) Feature plot of the subset of SSPCs, IIFCs, chondrocytes and osteoblasts from Figure 5. (Right) Outgoing and incoming interaction strengths of the subset of skeletal stem/progenitor cells (SSPCs), IIFCs, chondrocytes, and osteoblasts. (C) Dot plots of the expression of the ligands (underlined) and receptors of BMP, TGFβ, PDGF, POSTN, PTN, and ANGPTL family involved in cell–cell interactions from IIFCs after fracture.

Activation and differentiation trajectories of periosteal skeletal stem/progenitor cells (SSPCs) after fracture.

Periosteum-derived chondrocytes undergo cartilage to bone transformation.
A. UMAP projection of the subset of SSPCs, IIFCs, osteoblasts and chondrocytes in the integration of days 5 and 7 post-fracture datasets. B. Feature plots of Acan, Col10a1 and Ibsp expression. C. UMAP projection separated by time points. D. Percentage of cells in the hypertrophic/differentiating chondrocyte cluster.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (mice) | C57BL/6ScNj | Janvier Labs | ||
| Strain, strain background (mice) | B6.Cg-Tg(Prrx1-cre)1Cjt/J | Jackson Laboratory | IMSR_JAX:005584 | |
| Strain, strain background (mice) | B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J | Jackson Laboratory | IMSR_JAX:007676 | |
| Commercial assay or Kit | Chromium Single Cell Next GEM 3′ Library & Gel Bead Kit v.3.1 | 10X Genomics | ||
| Commercial assay or Kit | RNAscope Multiplex Fluorescent Assay V2 | Bio-Techne | ||
| Antibody | Rat monoclonal to mouse SCA1 | 740450, BD Biosciences | RRID:AB_2740177 | Dilution: 1/200 |
| Antibody | Rabbit monoclonal to mouse SOX9 | ab185230, Abcam | RRID:AB_2715497 | Dilution: 1/1000 |
| Antibody | Rabbit polyclonal to mouse Osterix/Sp7 | ab22552, Abcam | RRID:AB_2194492 | Dilution: 1/200 |
| Antibody | Goat polyclonal to mouse Periostin | AF2955, R&D Systems | RRID:AB_664123 | Dilution: 1/400 |
| Antibody | Goat polyclonal to mouse PECAM1 | AF3628, Bio-Techne | RRID:AB_2161028 | Dilution: 1/200 |
| Antibody | Rat monoclonal to mouse CD45 | 552848, BD Biosciences | RRID:AB_394489 | Dilution: 1/200 |
| Software, algorithm | Seurat | https://github.com/satijalab/seurat; Butler et al., 2024 | ||
| Software, algorithm | monocle3 | https://github.com/cole-trapnell-lab/monocle3; Pliner et al., 2024 | ||
| Software, algorithm | CytoTrace | https://cytotrace.stanford.edu/ | ||
| Software, algorithm | EnrichR | https://maayanlab.cloud/Enrichr/ | ||
| Software, algorithm | SCENIC | https://github.com/aertslab/SCENIC; Aibar, 2024 | ||
| Software, algorithm | STRING v11.5 database | https://string-db.org/ | ||
| Software, algorithm | CellChat | https://github.com/sqjin/CellChat; Jin, 2024 |
Additional files
-
Supplementary file 1
Lists of genes used for lineage score analyses of murine snRNAseq.
- https://cdn.elifesciences.org/articles/92519/elife-92519-supp1-v1.docx
-
Supplementary file 2
Lists of the regulons composing the cores.
- https://cdn.elifesciences.org/articles/92519/elife-92519-supp2-v1.docx
-
Supplementary file 3
Top 5 terms from Reactome analysis on fibro-core regulons.
- https://cdn.elifesciences.org/articles/92519/elife-92519-supp3-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/92519/elife-92519-mdarchecklist1-v1.docx