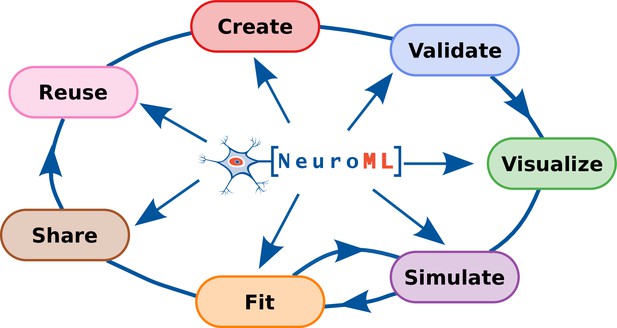
The NeuroML ecosystem for standardized multi-scale modeling in neuroscience
Figures
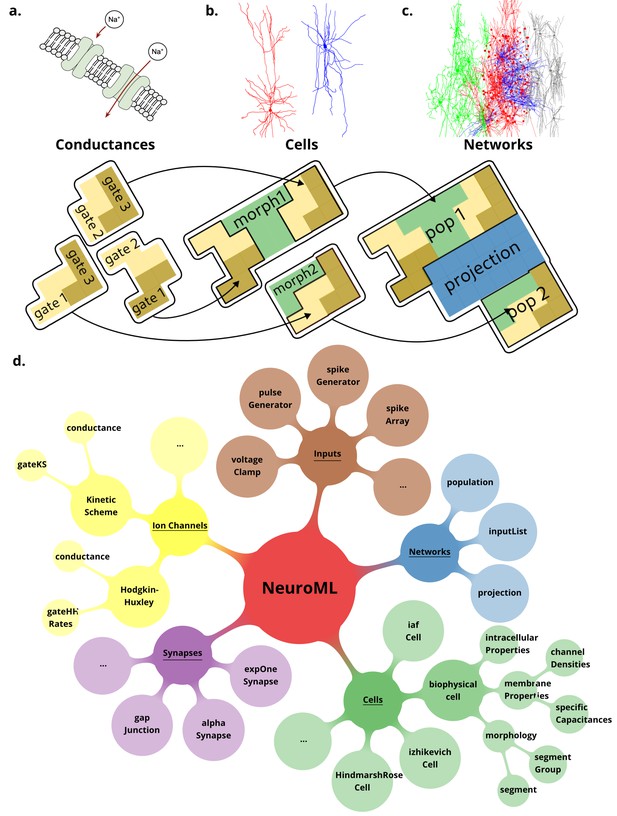
NeuroML is a modular, hierarchical format that supports multi-scale modeling.
Elements in NeuroML are formally defined, independent, self-contained building blocks with hierarchical relationships between them. (a) Models of ionic conductances can be defined as a composition of gates, each with specific voltage (and potentially [Ca2+]) dependence that controls the conductance. (b) Morphologically detailed neuronal models specify the 3D structure of the cells, along with passive electrical properties, and reference ion channels that confer membrane conductances. (c) Network models contain populations of these cells connected via synaptic projections. (d) A truncated illustration of the main categories of the NeuroMLv2 standard elements and their hierarchies. The standard includes commonly used model elements/building blocks that have been pre-defined for users: Cells: neuronal models ranging from simple spiking point neurons to biophysically detailed cells with multi-compartmental morphologies and active membrane conductances; Synapses and ionic conductance models: commonly used chemical and electrical synapse models (gap junctions), and multiple representations for ionic conductances; Inputs: to drive cell and network activity, e.g., current or voltage clamp, spiking background inputs; Networks: of populations (containing any of the aforementioned cell types), and projections. The full list of standard NeuroML elements can be found in Tables 1 and 2.
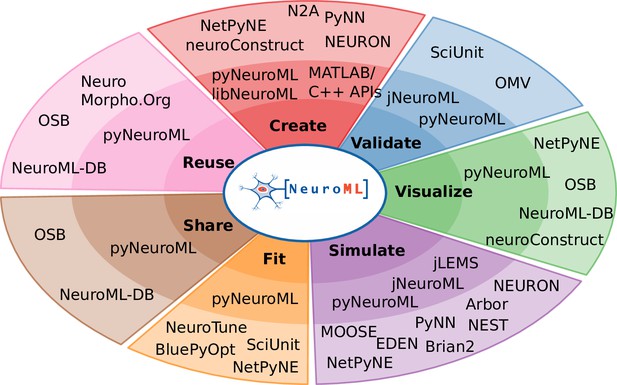
NeuroML compliant tools and their relation to the model life cycle.
The inner circle shows the core NeuroML tools and libraries that are maintained by the NeuroML developers. These provide the functionality to read, modify, or create new NeuroML models, as well as to validate, analyze, visualize and simulate the models. The outermost layer shows NeuroML-compliant tools that have been developed independently to allow various interactions with NeuroML models. These complement the core tools by facilitating model creation, validation, visualization, simulation, fitting/optimization, sharing, and reuse. Further information on each of the tools shown here can be found in Tables 3 and 4.
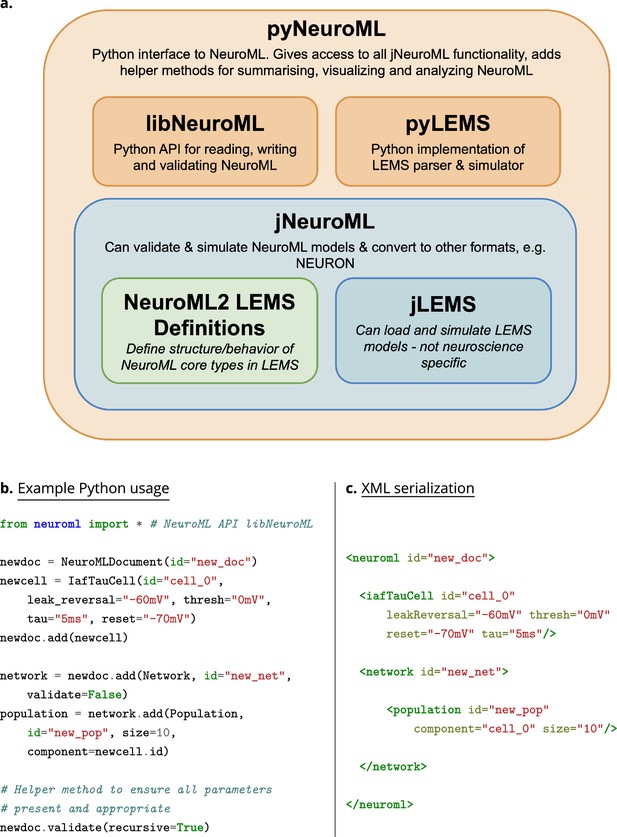
The core NeuroML software stack, and an example NeuroML model created using the Python NeuroML tools.
(a) The core NeuroML software stack consists of Java (blue) and Python (orange) based applications/libraries, and the LEMS model ComponentType definitions (green), wrapped up in a single package, pyNeuroML. Each of these modules can be used independently or the whole stack can be obtained by installing pyNeuroML with the default Python package manager, Pip: pip install pyneuroml. (b) An example of how to create a simple NeuroML model is shown, using the NeuroMLv2 Python API (libNeuroML) to describe a model consisting of a population of 10 integrate and fire point neurons (IafTauCell) in a network. The IafTauCell, Network, Population, and NeuroMLDocument model ComponentTypes are provided by the NeuroMLv2 standard. The underlying dynamics of the model are hidden from the user, being specified in the LEMS ComponentType definitions of the elements (see Methods). The simulator-independent NeuroML model description can be simulated on any of the supported simulation engines. (c) Extensible Markup Language (XML) serialization of the NeuroMLv2 model description shows the correspondence between the Python object model and the XML serialization.
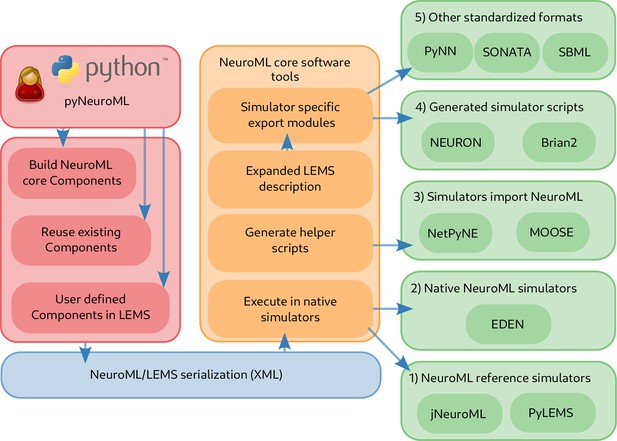
Workflow showing how to create and simulate NeuroML models using Python.
The Python API can be used to create models which may include elements built from scratch from the NeuroML standard, re-use elements from previously created models, or create new components based on novel model definitions expressed in LEMS (red). The generated model elements are saved in the default XML-based serialization (blue). The NeuroML core tools and libraries (orange) include modules to import model descriptions expressed in the XML serialization, and support multiple options for how simulators can execute these models (green). These include: (1) execution of the NeuroML models by reference simulators; (2) execution by other independently developed simulators that natively support NeuroML, such as EDEN; (3) generation of Python ‘import scripts’ which allow NeuroML models to be imported (and converted to internal formats) by simulators which support this; (4) fully expanding the LEMS description of the models, which can be mapped to generated simulator specific scripts for target simulators; (5) mapping to other standardized formats in neuroscience and systems biology.

PyNeuroML provides Python functions and command line utilities supporting all stages of the model life cycle.
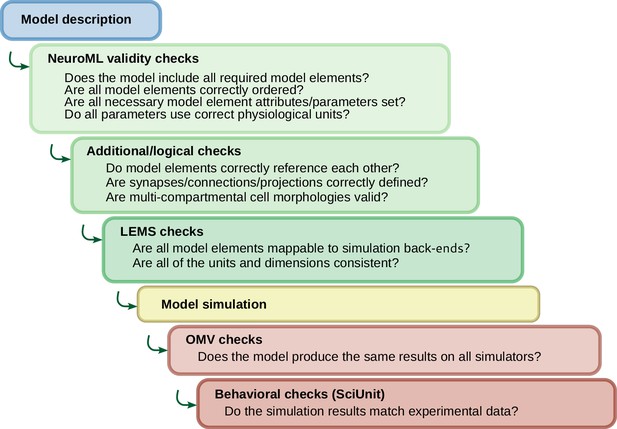
NeuroML model development incorporates multi-level validation of models.
Checks are performed on the model descriptions (blue) before simulation using validation at both the NeuroML and LEMS levels (green). After the models are simulated (yellow), further checks can be run to ensure the output is in line with expected behavior (brown). The OSB Model Validation (OMV) framework can be used to ensure consistent behavior across simulators, and comparisons can be made of model activity to their biological equivalents using SciUnit.
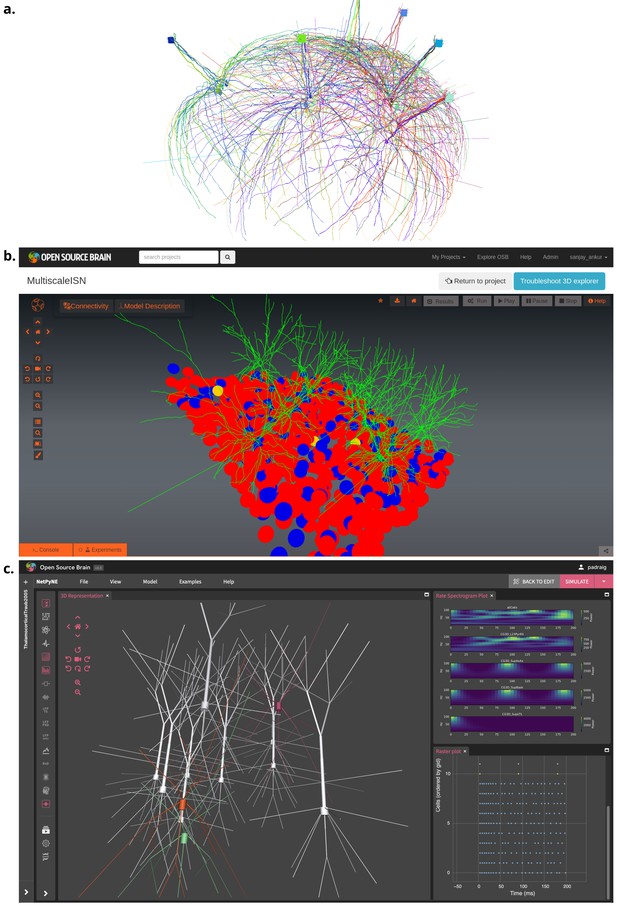
Visualization of detailed neuronal morphology of neurons and networks together with their functional properties (results from model simulation) enabled by NeuroML.
(a) Interactive 3-D (VisPy (Campagnola, 2023) based) visualization of an olfactory bulb network with detailed mitral and granule cells (Migliore et al., 2014), generated using pyNeuroML. (b) Visualization of an inhibition stabilized network based on Sadeh et al., 2017 using Open Source Brain (OSB) version 1 (Gleeson et al., 2019b). (c) Visualization of 3D network of simplified multi-compartmental cortical neurons (from Traub et al., 2005, imported as NeuroML Gleeson, 2019a) and simulated spiking activity using NetPyNE’s GUI (Dura-Bernal et al., 2019), which is embedded in OSB version 2.
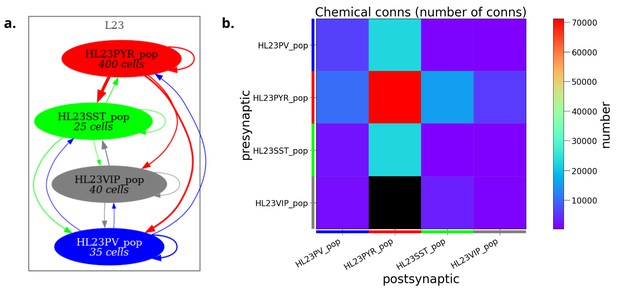
Analysis and visualization of network connectivity from NeuroML model descriptions prior to simulation.
Network connectivity schematic (a) and connectivity matrix (b) for a half scale implementation of the human layer 2/3 cortical network model (Yao et al., 2022) generated using pyNeuroML.
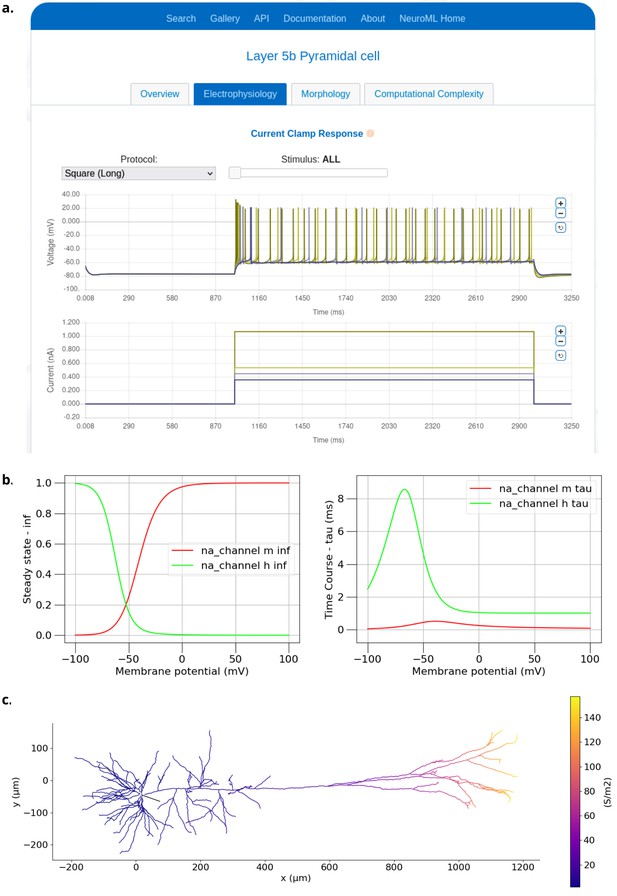
Examples of visualizing biophysical properties of a NeuroML model neuron.
(a) Electrophysiological properties generated by the NeuroML-DB web-based platform (Birgiolas et al., 2023). (Plots show four superimposed voltage traces in the top panel and corresponding current injection traces below). (b) Example plots of steady states of activation (na_channel na_m inf) and inactivation (na_channel na_h inf) variables and their time courses (na_channel na_m tau and na_channel na_h tau) for the Na channel from the classic Hodgkin Huxley model (Hodgkin and Huxley, 1952). (c) The distribution of the peak conductances for the Ih channel over a layer 5 Pyramidal cell (Hay et al., 2011). Both (b) and (c) were generated using the analysis features in pyNeuroML, and similar functionality is also available in OSBv1 (Gleeson et al., 2019b).
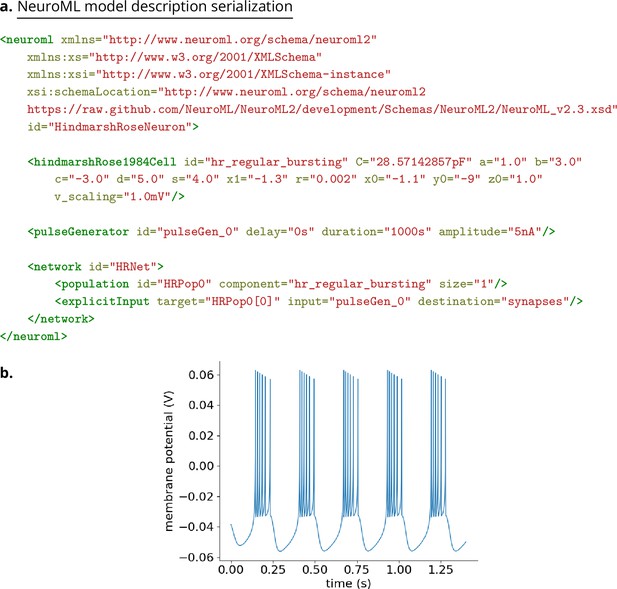
Example model description of a HindmarshRose1984Cell NeuroML component.
(a) XML serialization of the model description containing the main hindmarshRose1984Cell element with a set of parameters which result in regular bursting. A current clamp stimulus is applied using a pulseGenerator, and a population of one cell is added with this in a network. This XML can be validated against the NeuroML Schema. (b) Membrane potentials generated from a simulation of the model in (a). The LEMS simulation file to execute this is shown in Figure 15. The code used in this example is available here: https://github.com/OpenSourceBrain/HindmarshRose1984/tree/master/NeuroML2/examples.

Type definitions taken from the NeuroMLv2 schema (https://github.com/NeuroML/NeuroML2/blob/master/Schemas/NeuroML2/NeuroML_v2.3.1.xsd) which describes the structure of NeuroMLv2 elements.
Top: ‘simple’ types may not include other elements or attributes. Here, the Nml2Quantity_none and Nml2Quantity_voltage types define restrictions on the default string type to limit what strings can be used as valid values for attributes of these types. Bottom: example of a ‘complex’ type, the HindmarshRose cell model (Hindmarsh and Rose, 1984), that can also include other elements of other types, and extend other types.

LEMS ComponentType definition of the HindmarshRose cell model (Hindmarsh and Rose, 1984, https://github.com/NeuroML/NeuroML2/blob/master/NeuroML2CoreTypes/Cells.xml).

Example model description of a HindmarshRose1984Cell NeuroML component in Python using parameters for regular bursting.
This script generates the XML in Figure 11. The code used in this example is available here: https://github.com/OpenSourceBrain/HindmarshRose1984/tree/master/NeuroML2/examples.

An example simulation of the HindmarshRose model description shown in Figure 14 with the LEMS serialization shown at the bottom.
The code used in this example is available here: https://github.com/OpenSourceBrain/HindmarshRose1984/tree/master/NeuroML2/examples.
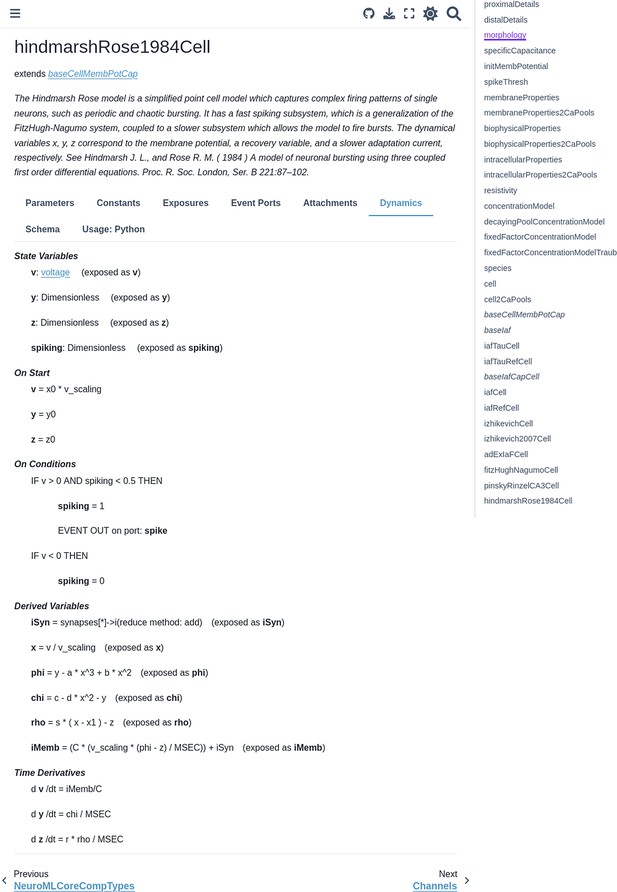
Documentation for the HindmarshRose1984Cell NeuroMLv2 ComponentType generated from the XSD schema and LEMS definitions on the NeuroML documentation website showing its dynamics (https://docs.neuroml.org/Userdocs/Schemas/Cells.html#hindmarshrose1984cell).
More information about the ComponentType can be obtained from the tabs provided.
Tables
Index of standard NeuroMLv2 ComponentTypes.
Index of standard NeuroMLv2 ComponentTypes (continued).
NeuroML software core tools and libraries, with a description of their scope, the main programming language they use (or other interaction means, e.g. Command Line Interface (CLI)), and links for more information.
| Tool | Language/interface | Description | URL |
|---|---|---|---|
| pyNeuroML | Python/CLI | Recommended Python library for NeuroML; provides pynml, primary command line tool for NeuroML | https://docs.neuroml.org/Userdocs/Software/pyNeuroML.html |
| libNeuroML | Python | Python API for NeuroML | https://docs.neuroml.org/Userdocs/Software/libNeuroML.html |
| NeuroMLlite | Python | High level library for creating NeuroML network models (beta) | https://docs.neuroml.org/Userdocs/Software/NeuroMLlite.html |
| PyLEMS | Python/CLI | Python API and simulator for LEMS | https://docs.neuroml.org/Userdocs/Software/pyLEMS.html |
| jLEMS | Java/CLI | Java API for LEMS and reference simulator | https://docs.neuroml.org/Userdocs/Software/jLEMS.html |
| org.neuroml.model | Java | Java API for NeuroML, DOI:10.5281/zenodo.5783290 | https://github.com/NeuroML/org.neuroml.model/ |
| org.neuroml.export | Java | Java API for translating NeuroML into different formats such as NEURON, DOI:10.5281/zenodo.1346272 | https://github.com/NeuroML/org.neuroml.export |
| org.neuroml.import | Java | Java API for importing formats into LEMS and NeuroML, DOI:10.5281/zenodo.5783295 | https://github.com/NeuroML/org.neuroml.import |
| jNeuroML | Java/CLI | Wraps jLEMS and all export/import packages and provides the jnml tool, DOI:10.5281/zenodo.593108 | https://docs.neuroml.org/Userdocs/Software/jNeuroML.html |
| NeuroML-C++ | C++ | C++ API for NeuroML | https://docs.neuroml.org/Userdocs/Software/NeuroML_API.html |
| NeuroML Toolbox | MATLAB | MATLAB NeuroML Toolbox | https://docs.neuroml.org/Userdocs/Software/MatLab.html |
Tools in the wi main programming language they use (or other interaction means, e.g. through a web browser, Graphical User Interface (GUI) or Command Line Interface (CLI)), and links for more information.
| Tool | Language/interface | Description | URL |
|---|---|---|---|
| Simulation engines | |||
| NEURON | Python/Hoc/CLI/GUI | Empirically-based simulations of neurons and networks of neurons | https://docs.neuroml.org/Userdocs/Software/Tools/NEURON.html |
| NetPyNE | Python/web | Package to facilitate the development, parallel simulation, analysis, and optimization of biological neuronal networks using the NEURON simulator. Also has a graphical web interface, NetPyNE-UI | https://docs.neuroml.org/Userdocs/Software/Tools/NetPyNE.html |
| EDEN | NeuroML | NeuroML-based neural simulator | https://docs.neuroml.org/Userdocs/Software/Tools/EDEN.html |
| MOOSE | Python | The Multiscale Object-Oriented Simulation Environment is the base and numerical core for large, detailed multi-scale simulations that span computational neuroscience and systems biology. Based on a reimplementation of the GENESIS 2 core. | https://docs.neuroml.org/Userdocs/Software/Tools/MOOSE.html |
| PyNN | Python | A simulator-independent language for building neuronal network models | https://docs.neuroml.org/Userdocs/Software/Tools/PyNN.html |
| NEST | Python/SLI | Simulator for spiking neural network models focusing on dynamics, size, and structure of neural systems | https://docs.neuroml.org/Userdocs/Software/Tools/NEST.html |
| Brian2 | Python | Easy to learn and use simulator for spiking neural networks | https://docs.neuroml.org/Userdocs/Software/Tools/Brian.html |
| Arbor | Python | A multi-compartment neuron simulation library | https://docs.neuroml.org/Userdocs/Software/Tools/Arbor.html |
| N2A | Java/GUI | Language and IDE for writing and simulating models | https://docs.neuroml.org/Userdocs/Software/Tools/N2A.html |
| Databases | |||
| OSB | Web | Resource for sharing and collaboratively developing computational models of neural systems | https://www.opensourcebrain.org/ |
| NeuroML-DB | Web | NeuroML database of cell and channel models | https://neuroml-db.org/ |
| Other tools | |||
| OMV | Python | Open Source Brain Model Validation framework | https://github.com/OpenSourceBrain/osb-model-validation |
| SciUnit | Python | Data driven unit testing framework | https://github.com/scidash/sciunit |
| BluePyOpt | Python | Blue Brain Python Optimization Library | https://bluepyopt.readthedocs.io/ |
| NeuroTune | Python | Package for fitting/optimization of NeuroML models | https://github.com/NeuralEnsemble/neurotune |
| PyElectro | Python | Electrophysiology analysis package | https://github.com/NeuralEnsemble/pyelectro |
Step-by-step guides for using NeuroML illustrating the various stages of the model development life cycle.
These include Introductory guides aimed at teaching the fundamental NeuroML concepts, Advanced guides illustrating specific modeling workflows, and Walkthrough guides discussing the steps required for converting models to NeuroML. An updated list is available at http://neuroml.org/gettingstarted.
| Link | Description | Model life cycle stages |
|---|---|---|
| Introductory guides | ||
| Guide 1 | Create and simulate a simple regular spiking Izhikevich neuron in NeuroML | Create, Validate, Simulate |
| Guide 2 | Create a network of two synaptically connected populations of Izhikevich neurons | Create, Validate, Visualize, Simulate |
| Guide 3 | Build and simulate a single compartment Hodgkin-Huxley neuron | Create, Validate, Visualize, Simulate |
| Guide 4 | Create and simulate a multi compartment hippocampal OLM neuron | Create, Validate, Visualize, Simulate |
| Advanced guides | ||
| Guide 5 | Create novel NeuroML models from components on NeuroML-DB | Reuse, Create, Validate, Simulate |
| Guide 6 | Optimize/fit NeuroML models to experimental data | Create, Validate, Simulate, Fit |
| Guide 7 | Extend NeuroML by creating a novel model type in LEMS | Create, Simulate |
| Walkthroughs | ||
| Guide 8 | Guide to converting cell models to NeuroML and sharing them on Open Source Brain | Create, Validate, Simulate, Share |
| Guide 9 | Conversion of Ray et al., 2020 | Create, Validate, Visualize, Simulate, Share |
Listing of validation tests run by NeuroML.
| Test | Description |
|---|---|
| Schema tests | |
| Check names | Check that names of all elements, attributes, parameters match those provided in the schema |
| Check types | Check that the types of all included elements |
| Check values | Check that values follow given restrictions |
| Check inclusion | Check that required elements are included |
| Check cardinality | Check the number of elements |
| Check hierarchy | Check that child/children elements are included in the correct parent elements |
| Check sequence order | Check that child/children elements are included in the correct order |
| Additional tests | |
| Check top level ids | Check that top level (root) elements have unique ids |
| Check Network level ids | Check that child/children of the Network element have unique ids |
| Check Cell Segment ids | Check that all Segments in a Cell have unique ids |
| Check single Segment without parent | Check that only one Segment is without parents (the soma Segment) |
| Check SegmentGroup ids | Check that all SegmentGroups in a Cell have unique ids |
| Check Member segment ids exist | Check that Segments referred to in SegmentGroup Members exist |
| Check SegmentGroup definition | Check that SegmentGroups being referenced are defined |
| Check SegmentGroup definition order | Check that SegmentGroups are defined before being referenced |
| Check included SegmentGroups | Check that SegmentGroups referenced by Include elements of other SegmentGroups exist |
| Check numberInternalDivisions | Check that SegmentGroups define numberInternalDivisions (used by simulators to discretize un-branched branches into compartments for simulation) |
| Check included model files | Check that model files included by other files exist |
| Check Population component | Check that a component id provided to a Population exists |
| Check ion channel exists | Check that an ion channel used to define a ChannelDensity element exists |
| Check concentration model species | Check that the species used in ConcentrationModel elements are defined |
| Check Population size | Check that the size attribute of a PopulationList matches the number of defined Instances |
| Check Projection component | Check that Populations used in the Projection elements exist |
| Check Connection Segment | Check that the Segment used in Connection elements exist |
| Check Connection pre/post cells | Check that the pre- and post-synaptic cells used in Connection elements exist and are correctly specified |
| Check Synapse | Check that the Synapse component used in a Projection element exists |
| Check root id | Check that the root Segment in a Cell morphology has id 0 |
Features supported by NeuroML in different simulation engines.
Note: the simulators themselves may support more features, but these have not been mapped onto by the NeuroML tools. Abstract cell models: abstract cell models included in the NeuroML standard (see Table 1). Single compartmental cells: neuronal models that include a single compartment (these engines do not support multi-compartmental cells). Multiple compartmental cells: neuronal models that include multiple compartments. Conductance-based models: models that support ionic conductances. Parallel execution: engines that support parallel execution using MPI/GPUs. Y: full support; N: no support; L: limited support in NeuroML toolchain.
| Tool | Abstract cell models | Single compartment cells | Multiple compartment cells | Conductance-based models | Parallel execution |
|---|---|---|---|---|---|
| jNeuroML/pyNeuroML | Y | Y | N | Y | N |
| NEURON | Y | Y | Y | Y | N |
| NetPyNE | Y | Y | Y | Y | Y |
| EDEN | Y | Y | Y | Y | Y |
| MOOSE | Y | Y | L | Y | N |
| PyNN | Y | Y | L | L | Y |
| NEST | Y | Y | N | N | Y |
| Brian2 | Y | Y | Y | Y | L |
| Arbor | L | Y | Y | L | Y |
Listing of NeuroML models and example repositories.
| Model | Description | URL |
|---|---|---|
| Neocortex | ||
| Billeh et al., 2020 | Morphologically detailed and point neuron models based on electrophysiological recordings from visual cortex neurons | https://github.com/OpenSourceBrain/AllenInstituteNeuroML |
| Brunel, 2000 | Spiking network illustrating balance between excitation and inhibition | https://github.com/OpenSourceBrain/Brunel2000 |
| Hay et al., 2011 | Layer 5 pyramidal cell model constrained by somatic and dendritic recordings | https://github.com/OpenSourceBrain/L5bPyrCellHayEtAl2011 |
| Izhikevich, 2004 | Spiking neuron model reproducing wide range of neuronal activity | https://github.com/OpenSourceBrain/IzhikevichModel |
| Markram et al., 2015 | Cell models from Neocortical Microcircuit of Blue Brain Project | https://github.com/OpenSourceBrain/BlueBrainProjectShowcase |
| Pospischil et al., 2008 | HH-based models for different classes of cortical and thalamic neurons | https://github.com/OpenSourceBrain/PospischilEtAl2008 |
| Potjans and Diesmann, 2014 | Microcircuit model of sensory cortex with 8 populations across 4 layers | https://github.com/OpenSourceBrain/PotjansDiesmann2014 |
| Dura-Bernal et al., 2017 | Model of mouse primary motor cortex (M1) | https://github.com/OpenSourceBrain/M1NetworkModel |
| Sadeh et al., 2017 | Point neuron model of Inhibition Stabilized Network | https://github.com/OpenSourceBrain/SadehEtAl2017-InhibitionStabilizedNetworks |
| Smith et al., 2013 | Layer 2/3 cell model used to investigate dendritic spikes | https://github.com/OpenSourceBrain/SmithEtAl2013-L23DendriticSpikes |
| Traub et al., 2005 | Single column network model containing 14 cell populations from cortex and thalamus | https://github.com/OpenSourceBrain/Thalamocortical |
| Bahl et al., 2012 | A set of reduced models of layer 5 pyramidal neurons | https://github.com/OpenSourceBrain/BahlEtAl2012_ReducedL5PyrCell |
| Wilson and Cowan, 1972 | A classic rate-based model describing the dynamics and interactions between the excitatory and inhibitory populations of neurons | https://github.com/OpenSourceBrain/WilsonCowan |
| Garcia Del Molino et al., 2017 | Rate-based model showing paradoxical response reversal of top-down modulation in cortical circuits with three interneuron types | https://github.com/OpenSourceBrain/del-Molino2017 |
| Mejias et al., 2016 | A rate-based model simulating the dynamics of a cortical laminar structure across multiple scales: intralaminar, interlaminar, interareal and whole cortex | https://github.com/OpenSourceBrain/MejiasEtAl2016 |
| Cerebellum | ||
| Maex and Schutter, 1998 | Cerebellar granule cell | https://github.com/OpenSourceBrain/GranuleCell |
| Cayco-Gajic et al., 2017 | Cerebellar granule cell layer network | https://github.com/SilverLabUCL/MF-GC-network-backprop-public |
| Maex and Schutter, 1998 | 3D Cerebellar granule cell layer network | https://github.com/OpenSourceBrain/GranCellLayer |
| Solinas et al., 2007 | Cerebellar Golgi cell model | https://github.com/OpenSourceBrain/SolinasEtAl-GolgiCell |
| Vervaeke et al., 2010 | Electrically connected cerebellar Golgi cell network model | https://github.com/OpenSourceBrain/VervaekeEtAl-GolgiCellNetwork |
| Hippocampus | ||
| Bezaire et al., 2016 | Full scale network model of CA1 region of hippocampus | https://github.com/mbezaire/ca1 |
| Ferguson et al., 2013 | Parvalbumin-positive interneuron from CA1, based on Izhikevich cell model | https://github.com/OpenSourceBrain/FergusonEtAl2013-PVFastFiringCell |
| Ferguson et al., 2014 | Pyramidal cell from CA1, based on Izhikevich cell model | https://github.com/OpenSourceBrain/FergusonEtAl2014-CA1PyrCell |
| Migliore et al., 2005 | Multi-compartmental model of pyramidal cell from CA1 region of hippocampus | https://github.com/OpenSourceBrain/CA1PyramidalCell |
| Pinsky and Rinzel, 1994 | Simplified model of CA3 pyramidal cell | https://github.com/OpenSourceBrain/PinskyRinzelModel |
| Wang and Buzsáki, 1996 | Hippocampal interneuronal network model exhibiting gamma oscillations | https://github.com/OpenSourceBrain/WangBuzsaki1996 |
| Olfactory bulb | ||
| Migliore et al., 2014 | Large-scale 3D olfactory bulb network with detailed mitral cells and granule cells | https://github.com/OpenSourceBrain/MiglioreEtAl14_OlfactoryBulb3D |
| Invertebrate | ||
| Hodgkin and Huxley, 1952 | Classic investigation of the ionic basis of the action potential | https://github.com/openworm/hodgkin_huxley_tutorial |
| FitzHugh, 1961 | Simplified form of Hodgkin Huxley model | https://github.com/OpenSourceBrain/FitzHugh-Nagumo |
| Prinz et al., 2004 | Pyloric network of the lobster stomatogastric ganglion system | https://github.com/OpenSourceBrain/PyloricNetwork |
| Boyle and Cohen, 2008 | Model of body wall muscle from C. elegans | https://github.com/openworm/muscle_model |
| Gleeson et al., 2018 | A multiscale framework for modeling the nervous system of C. elegans | https://github.com/openworm/c302 |
| General | ||
| Morris and Lecar, 1981 | Two dimensional reduced neuron model with calcium and potassium conductances | https://github.com/OpenSourceBrain/MorrisLecarModel |
| Hindmarsh and Rose, 1984 | A simplified point cell model which captures complex firing patterns of single neurons, such as periodic and chaotic bursting | https://github.com/OpenSourceBrain/HindmarshRose1984 |
| Showcases | ||
| NEST Showcase | Examples of interactions with simulator NEST | https://github.com/OpenSourceBrain/NESTShowcase |
| PyNN Showcase | Examples of interactions between NeuroML and PyNN | https://github.com/OpenSourceBrain/PyNNShowcase |
| NetPyNE Showcase | Examples of interactions between NeuroML and NetPyNE | https://github.com/OpenSourceBrain/NetPyNEShowcase |
| SBML Showcase | Examples of interactions between NeuroML and SBML | https://github.com/OpenSourceBrain/SBMLShowcase |
| Brian Showcase | Examples of interactions between NeuroML and Brian | https://github.com/OpenSourceBrain/BrianShowcase |
| MOOSE Showcase | Examples of interactions between NeuroML and MOOSE | https://github.com/OpenSourceBrain/MOOSEShowcase |
| Arbor Showcase | Examples of interactions between NeuroML and Arbor | https://github.com/OpenSourceBrain/ArborShowcase |
| EDEN Showcase | Examples of interactions between NeuroML and EDEN | https://github.com/OpenSourceBrain/EDENShowcase |
| The Virtual Brain Showcase | Examples of interactions between NeuroML and TVB | https://github.com/OpenSourceBrain/TheVirtualBrainShowcase |
| NEURON Showcase | Examples of interactions between NeuroML and NEURON | https://github.com/OpenSourceBrain/NEURONShowcase |
| neuroConstruct Showcase | Examples of neuroConstruct projects | https://github.com/OpenSourceBrain/neuroConstructShowcase |
| NeuroMorpho.Org | Examples of reconstructions from NeuroMorpho.Org | https://github.com/OpenSourceBrain/NeuroMorpho |
| Janelia MouseLight | Janelia MouseLight project neuronal reconstructions | https://github.com/OpenSourceBrain/MouseLightShowcase |