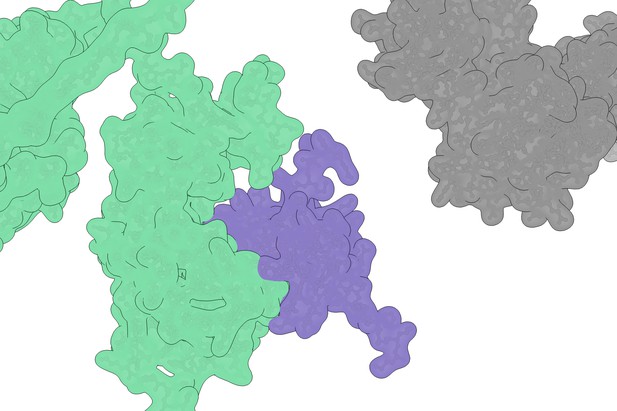
The protein K3 (purple) is depicted occupying the surface of the protein PKR (green), resulting in an inability to bind eIF2α (grey). Image credit: Chambers et al. (CC BY 4.0)
When viruses replicate, mutations can be introduced into their genetic information. These mutations can confer viruses with an advantage during infection: the proteins a virus uses to stick to host cells may change to bind better, allowing the virus to introduce itself into cells more easily; or the virus may lose a specific pattern in its surface, making it harder for the immune system to detect. These changes can make a virus more contagious or more deadly and can lead to the immune system acquiring mutations to counteract the viruses’ own.
In humans, a protein called PKR alerts host cells if a virus is present by adding a chemical tag to a second protein called eIF2α, which can halt the production of proteins in the cell, thus stopping viral replication. A type of virus, known as the poxvirus, has evolved a way to stop this ‘alarm’ by producing a protein called K3, which mimics eIF2α to intercept PKR and prevent the inhibition of protein synthesis. Both PKR and K3 seem to be evolving rapidly over time, gaining mutations that provide a competitive edge over one another. This is particularly interesting because PKR can adopt genetic changes that evade K3, but it can still recognize is natural substrate, eIF2α, which is not evolving.
The building blocks or amino acid residues in PKR that are rapidly evolving have been characterized. Additionally, several different versions or variants of PKR have been identified, where a change in a single residue allows K3 evasion, whilst maintaining its ability to halt protein synthesis.
Chambers et al. wanted to know how easily PKR can mutate to evade K3. The researchers used yeast to see how well variants of PKR functioned, since halting protein production impairs yeast growth. They screened 426 different versions of PKR, each with a change in a single building block, to see whether they retained the ability to target eIF2α, including when pitted against two variants of K3.
The results showed that PKR is extremely genetically pliable: it can change its surface to keep recognizing eIF2α while evading K3 in many different ways. Chambers et al. also characterized variants of PKR that were non-functional (because they could not bind eIF2α) and distinguished them from those that were susceptible to K3. They found that variants of PKR that could evade K3 could also evade an improved version of the protein. This suggests that the PKR variants that evade K3 are resilient.
Understanding this pattern of resiliency – both to mutations in PKR and to variants of K3 –may in the future aid therapeutic designs.




