Cholesterol taste avoidance in Drosophila melanogaster
Figures
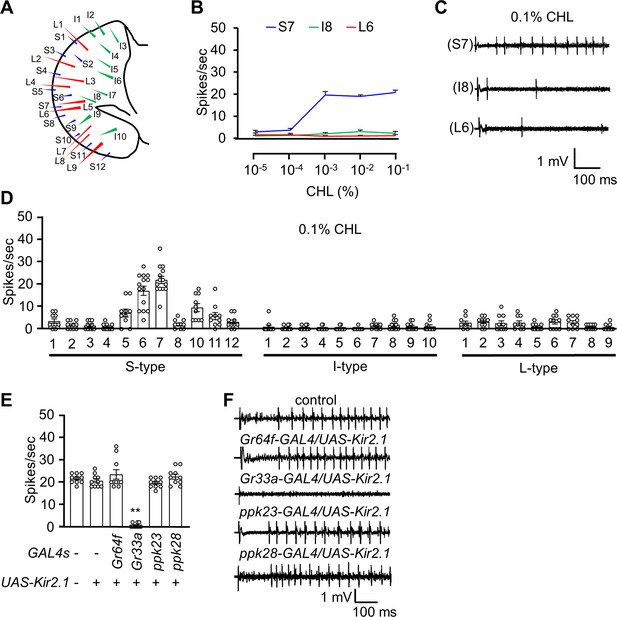
The neuronal response of the adult flies to cholesterol.
(A) Schematic diagram of the fly labellum. (B) Average frequencies of action potential generated from S7, I8, and L6 sensilla upon application of different concentrations of cholesterol (CHL; n=10–12). (C) Representative sample traces of S7, I8, and L6 from (B). (D) Electrophysiological responses of control flies produced from all labellum sensilla in response to 0.1% cholesterol (n=10–12). (E) Electrophysiological analysis of S7 sensilla in response to 0.1% cholesterol using flies in which different GRNs were inactivated by the inwardly rectifying potassium channel Kir2.1 (n=10–12). (F) Representative sample traces of the S7 sensilla from (E). All error bars represent SEMs. Single-factor ANOVA was combined with Scheffe’s post hoc analysis to compare multiple datasets. Asterisks indicate statistical significance compared to the control group (**p<0.01).
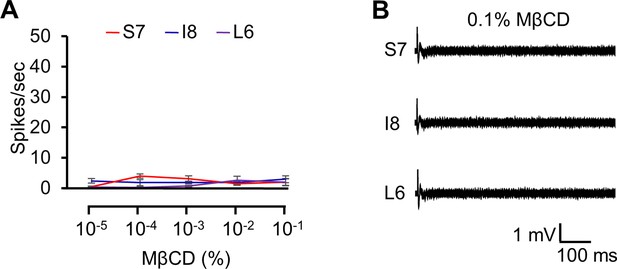
Electrophysiological responses using different doses of methyl-β-cyclodextrin (MβCD).
(A) Dose-dependent neuronal responses of w1118 adult flies to MβCD from S7, I8, and L6 sensilla (n=10). (B) Representative sample traces corresponding to the data in (A). Error bars represent standard errors of the means (SEMs). Statistical analysis was performed using single-factor ANOVA with Scheffe’s post hoc analysis to compare multiple datasets.
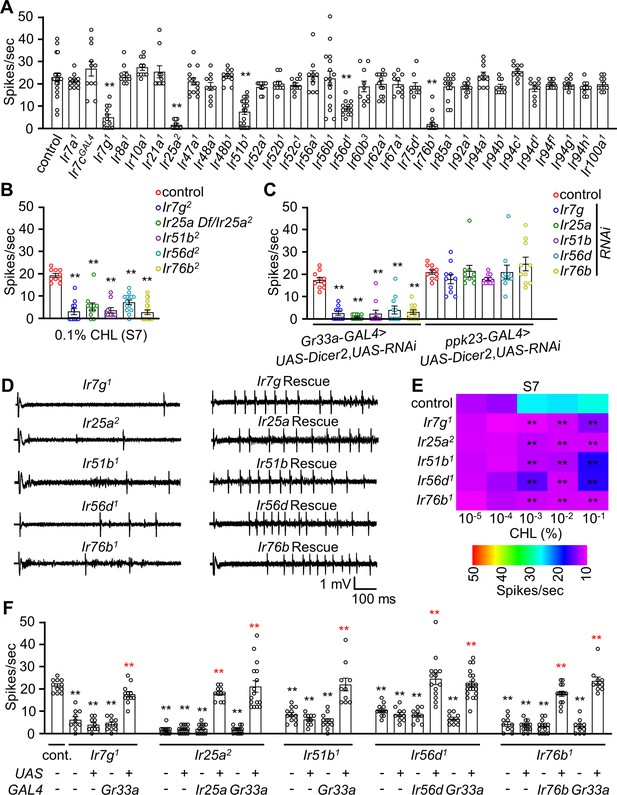
Ionotropic receptors (IRs) are responsible for sensing cholesterol.
(A) Tip recordings using 0.1% cholesterol to analyze the responses of S7 sensilla from control flies and from 32 Ir mutants (n=10–16). (B) Tip recordings using 0.1% cholesterol to analyze responses of S7 sensilla from Ir7g2, Ir25a Df/Ir25a2, Ir51b2, Ir56d2, and Ir76b2 (n=10–16). (C) Tip recordings using 0.1% cholesterol to analyze responses of S7 sensilla after RNAi knockdown of the following genes using either the Gr33a-GAL4 or ppk23-GAL4: Ir7g, Ir25a, Ir51b, Ir56d, and Ir76b. (D) Representative sample traces of (F) for control, mutants, and rescue lines using the GAL4/UAS system. (E) Heatmap representing the dose responses (spikes/sec) elicited by S7 sensilla from the control and the indicated mutants (Ir7g1, Ir25a2, Ir51b1, Ir56d1, and Ir76b1) (n=10–16). (F) Tip recordings performed on S7 sensilla (0.1% cholesterol) from control, Ir7g1, Ir25a2, Ir51b1, Ir56d1, Ir76b1, and from flies expressing the indicated cognate transgenes under control of either their own GAL4 or the Gr33a-GAL4 (n=10–14). All error bars represent SEMs. Single-factor ANOVA was combined with Scheffe’s post hoc analysis to compare multiple datasets. Black asterisks indicate statistical significance compared to the control group. The red asterisks indicate statistical significance between the control and the rescued flies (**p<0.01).
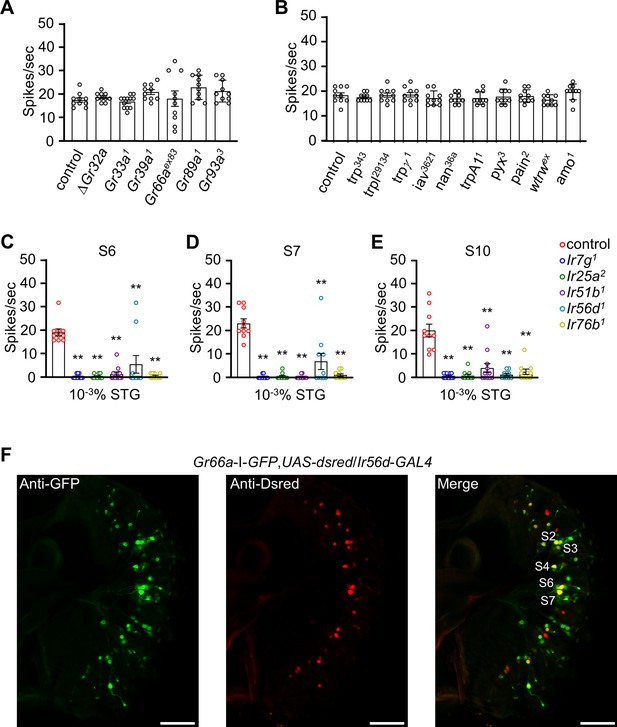
Electrophysiological analyses of S7 sensilla from mutants disrupting different bitter GRs andTRP channels in the presence of 10–1% CHL, and a subset of bitter GRNs express Ir56d .
(A) Tip recordings from S7 sensilla (using 0.1% cholesterol) from mutants disrupting broadly tuned bitter GRs (n=10). (B) Neuronal response analyses from S7 sensilla from trp mutant lines using 0.1% cholesterol (n=10). (C, D, E) Tip recording analyses of control flies and candidate Irs mutant flies (Ir7g1, Ir25a2, Ir51b1, Ir56d1, and Ir76b1) with 10–3% stigmasterol (STG) from S6, S7, and S10 sensilla (n=10–12). (F) Relative spatial distributions of the Gr66a (green; anti-GFP) and Ir56d (red; anti-DsRed) reporters in the labella of Gr66a-I-GFP, Ir56d-GAL4/UAS-DsRed flies. Images were acquired by confocal microscopy. The scale bars represent 50 µm. All error bars represent SEMs. Statistical analysis was performed using single-factor ANOVA with Scheffe’s post hoc analysis to compare multiple datasets. Asterisks indicate statistical significance compared to the control group (**p<0.01).
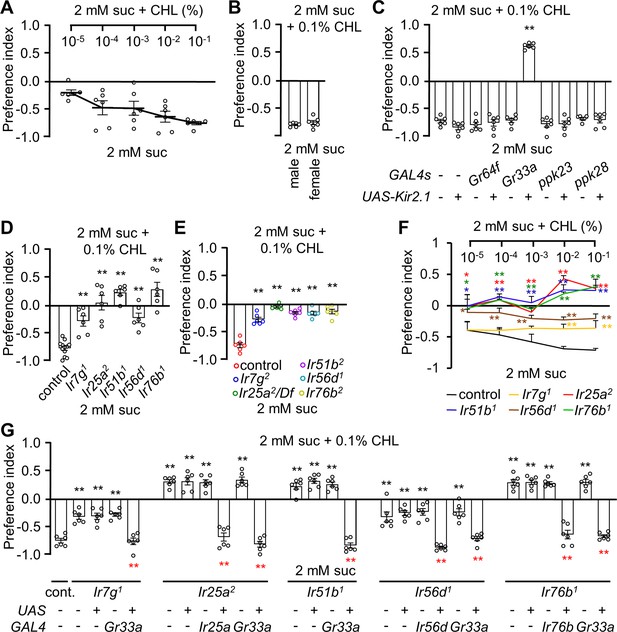
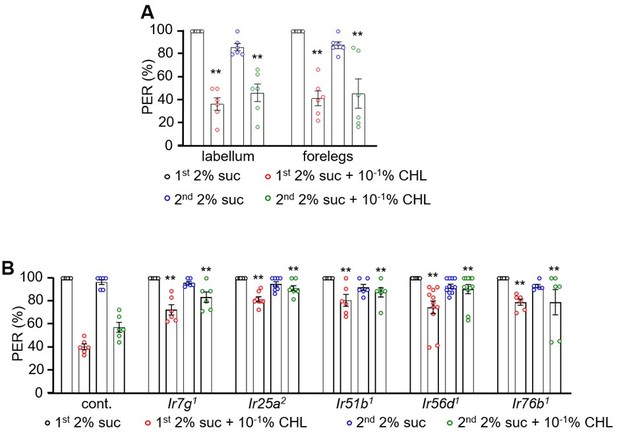
Ir7g, Ir25a, Ir51b, Ir56d, and Ir76b are required for the perception of cholesterol.
(A) Binary food choice analysis of w1118 adult flies toward different doses of cholesterol. Sucrose (2 mM) was included on both sides (n=6). (B) Binary food choice analyses to test for sex-specific difference in the feeding responses toward 0.1% cholesterol (n=6). (C) Binary food choice assays to determine the effects of inactivating different GRN types on the responses to 0.1% cholesterol. +/-indicates the presence or absence of the transgene, respectively (n=6). (D) Binary food choice assays to test the reponses of Ir7g1, Ir25a2, Ir51b1, Ir56d1, and Ir76b1 flies to 0.1% cholesterol (n=6). (E) Binary food choice assays to analyze the responses of Ir7g2, Ir25a Df, Ir51b2, Ir56d2, and Ir76b2 flies to 0.1% cholesterol (n=6). (F) Dose responses of control, Ir7g1, Ir25a2, Ir51b1, Ir56d1, and Ir76b1 flies to different concentrations of cholesterol (10–5%, 10–4%, 10–3%, 10–2%, and 10–1%) via binary food choice assays (n=6). (G) Rescue of Ir7g1, Ir25a2, Ir51b1, Ir56d1, and Ir76b1 defects by expressing the wild-type cDNAs under the control of the GAL4 drivers specific to each gene (Ir25a, Ir56d, and Ir76b) or Gr33a-GAL4 (n=6). All error bars represent SEMs. Single-factor ANOVA was combined with Scheffe’s post hoc analysis to compare multiple datasets. Black asterisks indicate statistical significance compared to the control group. The red asterisks indicate statistical significance between the control and the rescued flies (**p<0.01).
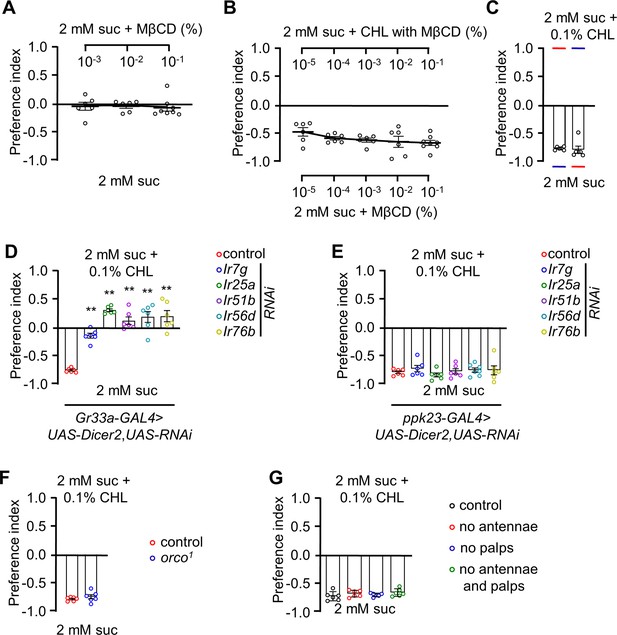
Binary food choice assays with CHL and methyl-β-cyclodextrin (MβCD).
(A) Dose-dependent binary food choice assays using control flies with 10–3%, 10–2%, and 10–1% MβCD containing 2 mM sucrose vs 2 mM sucrose only (n=6). (B) Dose-dependent binary food choice assay comparing cholesterol (CHL) vs MβCD food. Sucrose (2 mM) was employed on both sides (n=6). (C) Behavioral analysis of control flies after switching the red and blue dyes in the two food options (n=6). (D) Binary food choice assays using flies expressing UAS-RNAi lines for Ir7g, Ir25a, Ir51b, Ir56d, and Ir76b with combined with UAS-Dicer2 and driven by the Gr33a-GAL4. (E) Binary food choice assays using flies expressing UAS-RNAi lines for Ir7g, Ir25a, Ir51b, Ir56d, and Ir76b combined with UAS-Dicer2 and driven by the ppk23-GAL4 (n=6). (F) Binary food choice assays using control flies and orco1 mutants (n=6). (G) Evaluation of the role of olfactory organs in rejecting 0.1% cholesterol using binary food choice assays (n=6). All error bars represent SEMs. Statistical analysis was performed using single-factor ANOVA with Scheffe’s post hoc analysis to compare multiple datasets. Asterisks indicate statistical significance compared to the control group (**P<0.01).
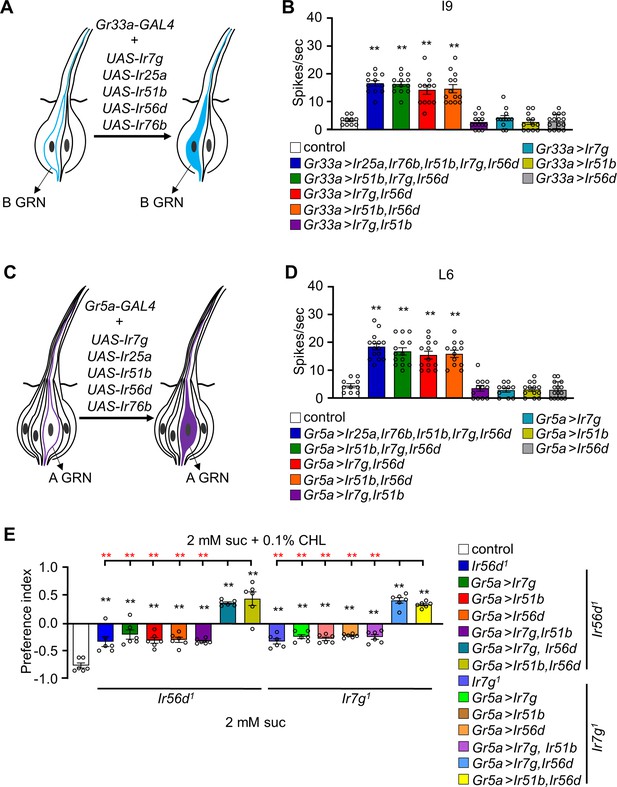
Testing whether ectopic expression of Ir7g, Ir25a, Ir51b, Ir56d, and Ir76b in L- and I-type sensilla confers cholesterol responsiveness.
(A) Schematic representation of ectopic expression of Irs in B GRNs under control of the Gr33a-GAL4. (B) Tip recordings conducted from I9 sensilla with 0.1% cholesterol using flies overexpressing UAS-Ir7g, UAS-Ir25a, UAS-Ir51b, UAS-Ir56d, and UAS-Ir76b in B GRNs under control of the Gr33a-GAL4 (n=10–16). (C) Schematic presentation of misexpression of Irs in A GRNs under control of the Gr5a-GAL4. (D) Tip recordings from L6 sensilla of the indicated flies expressing the indicated Irs under control of the Gr5a-GAL4 (n=10–16). (E) Binary food choice assays testing for attraction or aversion to 0.1% cholesterol in flies misexpressing Ir7g, Ir51b, and Ir56d in A GRNs (Gr5a-GAL4). The Irs were ectopically expressed in either an Ir56d1 or Ir7g1 mutant background (n=6). The red asterisks indicate the comparison of the combination of two UAS lines (Ir7g, Ir56d and Ir51b, Ir56d) driven by Gr5a-GAL4 with all the single UAS line including the combination of Ir7g and Ir51b. All error bars represent SEMs. Single-factor ANOVA was combined with Scheffe’s post hoc analysis to compare multiple datasets. Black asterisks indicate statistical significance compared with the control (**p<0.01).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Drosophila melanogaster) | Ir7a1 | Rimal et al., 2019 | Provided by Dr. Y. Lee | |
| Genetic reagent (Drosophila melanogaster) | Ir7g1: y1 w* Mi{y+mDint2=MIC}Ir7gMI06687 | Bloomington Drosophila Stock Center | BDSC:42420 | |
| Genetic reagent (Drosophila melanogaster) | Ir8a1:w[*]TI{w[+m*] =TI}Ir8a(1);Bl(1)L(2)/CyO | Bloomington Drosophila Stock Center | BDSC:23842 | |
| Genetic reagent (Drosophila melanogaster) | Ir10a1:w1118 Mi{GFPE.3xP3=ET1}Ir10aMB03273 | Bloomington Drosophila Stock Center | BDSC:41744 | |
| Genetic reagent (Drosophila melanogaster) | Ir21a1: w1118; PBac{w+mC=PB}Ir21ac02720 | Bloomington Drosophila Stock Center | BDSC:10975 | Provided by Dr. C. Montell |
| Genetic reagent (Drosophila melanogaster) | Ir25a2 | Benton et al., 2009 | Provided by Dr. L. Voshall | |
| Genetic reagent (Drosophila melanogaster) | Ir47a1 | Rimal et al., 2019 | Provided by Dr. Y. Lee | |
| Genetic reagent (Drosophila melanogaster) | Ir48a1: w1118; Mi{GFPE.3xP3 =ET1}Ir48aMB09217 | Bloomington Drosophila Stock Center | BDSC:26453 | |
| Genetic reagent (Drosophila melanogaster) | Ir48b1:w1118;Mi{GFPE.3xP3=ET1}Ir48bMB02315 | Bloomington Drosophila Stock Center | BDSC:23473 | |
| Genetic reagent (Drosophila melanogaster) | Ir51b1:w1118;PBac{w+mC=PB}rowc00387 Ir51bc00387 | Bloomington Drosophila Stock Center | BDSC:10046 | |
| Genetic reagent (Drosophila melanogaster) | Ir52a1 | Rimal et al., 2019 | Provided by Dr. Y. Lee | |
| Genetic reagent (Drosophila melanogaster) | Ir52b1:w1118;Mi{GFPE.3xP3 =ET1}Ir52bMB02231/SM6a | Bloomington Drosophila Stock center | BDSC:25212 | |
| Genetic reagent (Drosophila melanogaster) | Ir52c1:w1118; Mi{GFPE.3xP3 =ET1}Ir52cMB04402 | Bloomington Drosophila Stock center | BDSC:24580 | |
| Genetic reagent (Drosophila melanogaster) | Ir56a1 | Rimal et al., 2019 | Provided by Dr. Y. Lee | |
| Genetic reagent (Drosophila melanogaster) | Ir56b1:w1118;Mi{GFPE.3xP3 =ET1}Ir56bMB09950 | Bloomington Drosophila Stock Center | BDSC:27818 | |
| Genetic reagent (Drosophila melanogaster) | Ir56d1:w[*];Ir56d1 | Bloomington Drosophila Stock Center | BDSC:81249 | |
| Genetic reagent (Drosophila melanogaster) | Ir60b3 | Sang et al., 2024 | Provided by Dr. Y. Lee | |
| Genetic reagent (Drosophila melanogaster) | Ir62a1:y1w*;Mi{y+mDint2=MIC}Ir62aMI00895 Iml1MI00895/TM3, Sb1 Ser1 | Bloomington Drosophila Stock Center | BDSC:32713 | |
| Genetic reagent (Drosophila melanogaster) | Ir67a1: y1 w*; Mi{y+mDint2 =MIC}Ir67aMI11288 | Bloomington Drosophila Stock Center | BDSC:56583 | |
| Genetic reagent (Drosophila melanogaster) | Ir75d1:w1118;Mi{GFPE.3xP3 =ET1}Ir75dMB04616 | Bloomington Drosophila Stock Center | BDSC:24205 | |
| Genetic reagent (Drosophila melanogaster) | Ir76b1 | Zhang et al., 2013a | Provided by Dr. C. Montell | |
| Genetic reagent (Drosophila melanogaster) | Ir85a1:w1118;Mi{GFPE.3xP3=ET1}Ir85aMB04613 Pif1AMB04613 | Bloomington Drosophila Stock Center | BDSC:24590 | |
| Genetic reagent (Drosophila melanogaster) | Ir92a1:w1118;Mi{GFPE.3xP3=ET1}Ir92aMB03705 | Bloomington Drosophila Stock Center | BDSC:23638 | |
| Genetic reagent (Drosophila melanogaster) | Ir94a1 | Rimal et al., 2019 | Provided by Dr. Y. Lee | |
| Genetic reagent (Drosophila melanogaster) | Ir94b1:w1118; Mi{GFPE.3xP3=ET1}Ir94bMB02190 | Bloomington Drosophila Stock Center | BDSC:23424 | |
| Genetic reagent (Drosophila melanogaster) | Ir94c1 | Rimal et al., 2019 | Provided by Dr. Y. Lee | |
| Genetic reagent (Drosophila melanogaster) | Ir94d1:y1w[;Mi{y+mDint2=MIC} Ir94dMI01659CG17380MI01659 | Bloomington Drosophila Stock Center | BDSC:33132 | |
| Genetic reagent (Drosophila melanogaster) | Ir94f1: y1 w*; Mi{y+mDint2=MIC}Ir94fMI00928 | Bloomington Drosophila Stock Center | BDSC:33095 | |
| Genetic reagent (Drosophila melanogaster) | Ir94g1: w1118; Mi{GFPE.3xP3=ET1}Ir94gMB07445 | Bloomington Drosophila Stock Center | BDSC:25551 | |
| Genetic reagent (Drosophila melanogaster) | Ir94h1 | Rimal et al., 2019 | Provided by Dr. Y. Lee | |
| Genetic reagent (Drosophila melanogaster) | Ir100a1: w1118;P{w+mC=EP}Ir100aG19846 CG42233G19846 | Bloomington Drosophila Stock Center | BDSC:31853 | |
| Genetic reagent (Drosophila melanogaster) | UAS-Ir25a | Lee et al., 2018 | Provided by Dr. Y. Lee | |
| Genetic reagent (Drosophila melanogaster) | UAS-Ir51b | Dhakal et al., 2021 | Provided by Dr. Y. Lee | |
| Genetic reagent (Drosophila melanogaster) | Gr33a1 | Moon et al., 2009 | Provided by Dr. C. Montell | |
| Genetic reagent (Drosophila melanogaster) | Gr33a-GAL4 | Moon et al., 2009 | Provided by Dr. C. Montell | |
| Genetic reagent (Drosophila melanogaster) | Gr47a1 | Lee et al., 2015 | Provided by Dr. C. Montell | |
| Genetic reagent (Drosophila melanogaster) | elav-GAL4;UAS-Dicer2 | Bloomington Drosophila Stock Center | BDSC:25750 | |
| Genetic reagent (Drosophila melanogaster) | Gr39a1 | Bloomington Drosophila Stock Center | BDSC:10562 | |
| Genetic reagent (Drosophila melanogaster) | Gr93a3 | Lee et al., 2009 | Provided by Dr. Y. Lee | |
| Genetic reagent (Drosophila melanogaster) | UAS-Kir2.1 | Bloomington Drosophila Stock Center | BDSC:6596 | |
| Genetic reagent (Drosophila melanogaster) | ΔGr32a | Miyamoto and Amrein, 2008 | Provided by Dr. H. Amrein | |
| Genetic reagent (Drosophila melanogaster) | Gr66aex83 | Moon et al., 2006 | Provided by Dr. C. Montell | |
| Genetic reagent (Drosophila melanogaster) | Gr89a1 | Korea Drosophila Resource Center | KDRC: (Sung et al., 2017) | |
| Genetic reagent (Drosophila melanogaster) | Ir7cGAL4 | McDowell et al., 2022 | Provided by Dr. M. Gordon | |
| Genetic reagent (Drosophila melanogaster) | Ir20a1 | Ganguly et al., 2017 | Provided by Dr. A. Dahanukar | |
| Genetic reagent (Drosophila melanogaster) | Ir25a-GAL4 | Benton et al., 2009 | Provided by Dr. L. Vosshall | |
| Genetic reagent (Drosophila melanogaster) | UAS-Ir76b | Moon et al., 2006 | Provided by Dr. C. Montell | |
| Genetic reagent (Drosophila melanogaster) | Ir76b-GAL4 | Moon et al., 2006 | Provided by Dr. C. Montell | |
| Genetic reagent (Drosophila melanogaster) | ppk23-GAL4 | Thistle et al., 2012 | Provided by Dr. K. Scott | |
| Genetic reagent (Drosophila melanogaster) | ppk28-GAL4 | Cameron et al., 2010 | Provided by Dr. H. Amrein | |
| Genetic reagent (Drosophila melanogaster) | Gr5a-GAL4 | Dahanukar et al., 2001 | Provided by Dr. H. Amrein | |
| Genetic reagent (Drosophila melanogaster) | UAS-Kir2.1 | Bloomington Drosophila Stock Center | BDSC:6595 | |
| Genetic reagent (Drosophila melanogaster) | Ir7g2 | Pradhan et al., 2024 | Provided by Dr. Y. Lee | |
| Genetic reagent (Drosophila melanogaster) | UAS-Ir7g | Pradhan et al., 2024 | Provided by Dr. Y. Lee | |
| Genetic reagent (Drosophila melanogaster) | UAS-Ir56d | Sánchez-Alcañiz et al., 2018 | Provided by Dr. R. Benton | |
| Genetic reagent (Drosophila melanogaster) | Ir56d-GAL4 | Korea Drosophila Resource Center | KDRC:2307 | |
| Genetic reagent (Drosophila melanogaster) | Ir56d2 | Bloomington Drosophila Stock Center | BDSC:81250 | |
| Genetic reagent (Drosophila melanogaster) | Ir51b2 | Dhakal et al., 2021 | Provided by Dr. Y. Lee | |
| Genetic reagent (Drosophila melanogaster) | BC/CyO;Gr66a-I-GFP,UAS-dsred/TM6b | Weiss et al., 2011 | Provided by Dr. J.R. Carlson | |
| Genetic reagent (Drosophila melanogaster) | Ir7g RNAi | Vienna Drosophila Resource Center | VDRC:100885 | |
| Genetic reagent (Drosophila melanogaster) | Ir25a RNAi | Vienna Drosophila Resource Center | VDRC:106731 | |
| Genetic reagent (Drosophila melanogaster) | Ir51b RNAi | Vienna Drosophila Resource Center | VDRC:29984 | |
| Genetic reagent (Drosophila melanogaster) | Ir56d RNAi | Vienna Drosophila Resource Center | VDRC6112 | |
| Genetic reagent (Drosophila melanogaster) | Ir76b RNAi | Vienna Drosophila Resource Center | VDRC8433 | |
| Genetic reagent (Drosophila melanogaster) | trpA11 | Kwon et al., 2008 | Provided by Dr. C. Montell | |
| Genetic reagent (Drosophila melanogaster) | trpl29134 | Niemeyer et al., 1996 | Provided by Dr. C. Montell | |
| Genetic reagent (Drosophila melanogaster) | trpγ1 | Akitake et al., 2015 | Provided by Dr. C. Montell | |
| Genetic reagent (Drosophila melanogaster) | amo1 | Watnick et al., 2003 | Provided by Dr. C. Montell | |
| Genetic reagent (Drosophila melanogaster) | iav3621 | Bloomington Drosophila Stock center | BDSC:24768 | |
| Genetic reagent (Drosophila melanogaster) | nan36a | Kim et al., 2003 | Provided by Dr. C. Kim | |
| Genetic reagent (Drosophila melanogaster) | trp343 | Tracey et al., 2003 | Provided by Dr. C. Montell | |
| Genetic reagent (Drosophila melanogaster) | pyx3 | Lee et al., 2005 | Provided by Dr. Y. Lee | |
| Genetic reagent (Drosophila melanogaster) | wtrwex | Kim et al., 2010 | Provided by Dr. C. Montell | |
| Genetic reagent (Drosophila melanogaster) | pain2 | Tracey et al., 2003 | Provided by Dr. S. Benzer | |
| Antibody | Rabbit anti-DsRed(rabbit polyclonal) | Takara | Cat # 632496 RRID:AB_10013483 | 1:1000 (1 µL) |
| Antibody | Goat anti-mouse Alexa Fluor 568 | Thermo fisher/Invitrogen | Cat # A11004 RRID:AB_2534072 | 1:200 (1 µL) |
| Antibody | Mouse anti-GFP (mouse monoclonal) | Molecular probe | Cat # A11120 RRID:AB_221568 | 1:1000 (1 µL) |
| Antibody | Goat anti-mouse Alexa Fluor 488 | Thermo Fisher/Invitrogen | Cat # A11029 RRID:AB_2534088 | 1:200 (1 µL) |
| Chemical compound or drug | Cholesterol | Sigma-Aldrich Co. | Cat# C4951 | |
| Chemical compound or drug | Sucrose | Sigma-Aldrich Co. | Cat# S9378 | |
| Chemical compound or drug | Tricholine citrate | Sigma-Aldrich Co. | Cat# T0252 | |
| Chemical compound or drug | Stigmasterol | Sigma-Aldrich Co. | Cat# S2424 | |
| Chemical compound or drug | Sulforhodamine B | Sigma-Aldrich Co. | Cat# 230162 | |
| Chemical compound or drug | Brilliant blue FCF | Wako Pure Chemical Industry Ltd. | Cat# 027–12842 | |
| Chemical compound or drug | Methyl beta cyclodextrin | Sigma-Aldrich Co. | Cat# 332615 | |
| Chemical compound or drug | Paraformaldehyde | Electron Microscopy Sciences | Cat # 15710 | 1:500 Provided by Dr. J.A. Veenstra |
| Chemical compound or drug | Goat Serum, New Zealand origin | Gibco | Cat # 16210064 | |
| Software, algorithm | Origin Pro Version | OriginLab corporation | RRID:SCR_002815 | https://www.originlab.com/ |
| Software, algorithm | Graphpad Prism | GraphPad | RRID:SCR_002798 | https://www.graphpd.com/ |
| Software, algorithm | Autospike 3.1 software | https://www.syntech.co.za/ |
Additional files
-
Supplementary file 1
Electrophysiological analysis and binary food choice assay of MβCD, cholesterol, and stigmasterol, and immunohistochemical analysis of Ir56d-GAL4 co-localization with a bitter GRN reporter were performed.
- https://cdn.elifesciences.org/articles/106256/elife-106256-supp1-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/106256/elife-106256-mdarchecklist1-v1.docx