A gene-specific T2A-GAL4 library for Drosophila
Figures
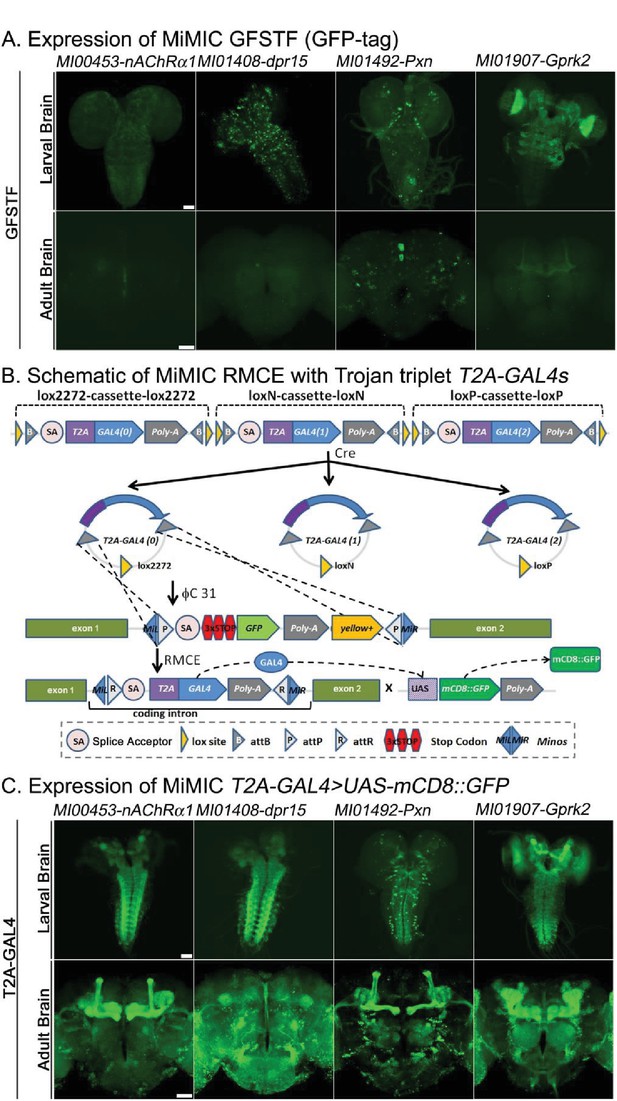
Protein distribution and expression patterns of genes containing MiMICs tagged with GFSTF or T2A-GAL4.
The MiMIC transposon contains two inverted attP sites that allow RMCE. (A) Detection of the expression domains of the indicated genes tagged with GFSTF in larvae and adult brains. GFP: green (B) Schematic of the MiMIC conversion with Trojan triplet T2A-GAL4 cassettes (Diao et al., 2015). Only the inserted T2A-GAL4 cassette with the correct orientation and phase results in GAL4 expression that drives UAS-mCD8::GFP expression. (C) Detection of the expression domains in larvae and adult brains of genes tagged with T2A-GAL4 using UAS-mCD8::GFP. mCD8::GFP: green. Scale bar: 50 μm.
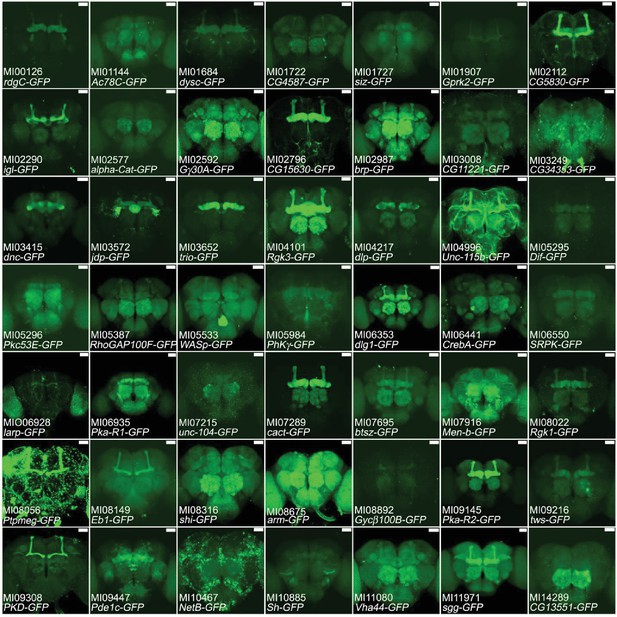
Expression of MiMIC GFSTFs tagged genes in adult brains.
We document the expression patterns and protein localization associated with 49 genes. The MiMIC identification number and affected genes are shown. Note that only ~19% of all genes tested showed detectable GFP signals in adult brains. Images are available on Flypush (http://flypush.imgen.bcm.tmc.edu/pscreen/rmce/). Many patterns are in agreement with the expression patterns of published genes/proteins (e.g. brp, Rgk1, dlg1, cact). Green: GFSTF. Scale bar: 50 μm.
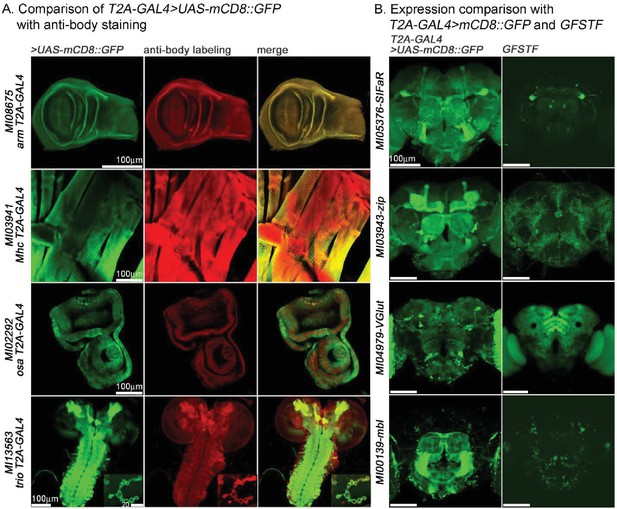
Similarities and differences between expression patterns associated with GAL4 >UAS GFP driven patterns and endogenous proteins in adult brains.
(A) Comparison of T2A-GAL4 > UAS-mCD8::GFP with anti-body stainings for arm (larval wing disc), Mhc (larval muscle), osa (larval eye-antennal imaginal disc) and trio (larval CNS and boutons/NMJ). Green: mCD8::GFP, Red: antibody labeling. (B) Comparison of T2A-GAL4 > UAS-mCD8::GFP with GFSTF for SIFaR, zip, VGlut and mbl in adult brains. Note the striking differences. Green in left panel: mCD8::GFP; Green in right panel: GFSTF. Scale bar is shown in figure.
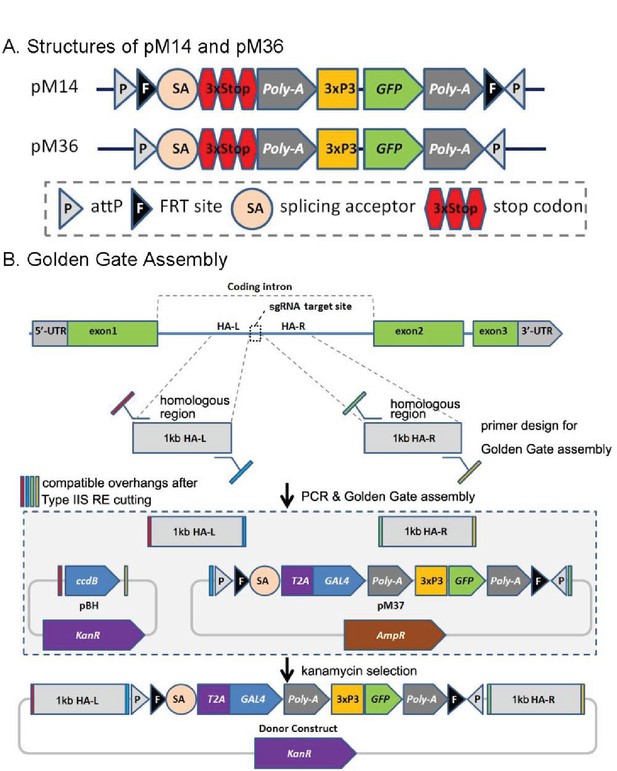
CRIMIC pM14 and pM36, and Golden Gate Assembly.
(A) Structures of pM14 and pM36. The CRIMIC pM14 cassette contains MiMIC-like cassette (SA-3xstop-polyA) and two FRT sites. The CRIMIC pM36 cassette was modified by removing the two FRT sites from PM14. (B) Golden Gate Assembly. Two sets of primers containing Type IIS RE sites are typically used to amplify ~1 kb homology arms by PCR. These arms, pM37 DNA and pBH vector (KanR) digested with Type IIS Restriction Enzymes and cloned using Golden Gate Assembly to generate the donor construct in a single reaction. The pM14/pM36 based donor DNAs were constructed with the same approach. The complete donor construct is selected with kanamycin. The components in these diagrams are not drawn to scale.
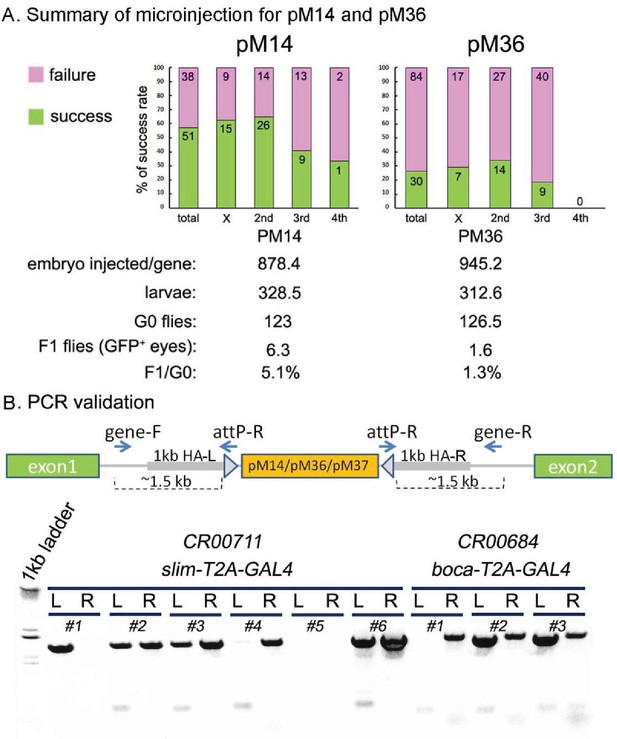
The efficiency of cassette insertion with CRIMIC pM14 and pM36, and PCR validation.
(A) Summary of the microinjection success rates for pM14 and pM36. We injected embryos for 89 genes with pM14 and the success rate was 57.3%. We also injected embryos for 114 genes with pM36 and the success rate was 26.3%. (B) The CRIMIC flies were verified by PCR to ensure that cassettes are in the correct position. Two specific primers outside homology arms (gene-F/gene-R) in each gene were designed to amplify the homologous arm regions by PCR with attP-R primer. The CR00711 (slim T2A-GAL4) and CR00684 (boca T2A-GAL4) are shown here. The genomic DNA of individual alleles was extracted and PCR was performed (see Materials and methods). The #2, #3 and #6 of slim T2A-GAL4s are correct alleles, and #2 and #3 of boca T2A-GAL4s are correct alleles. L: left region of the inserted cassette; R: right region of the inserted cassette.
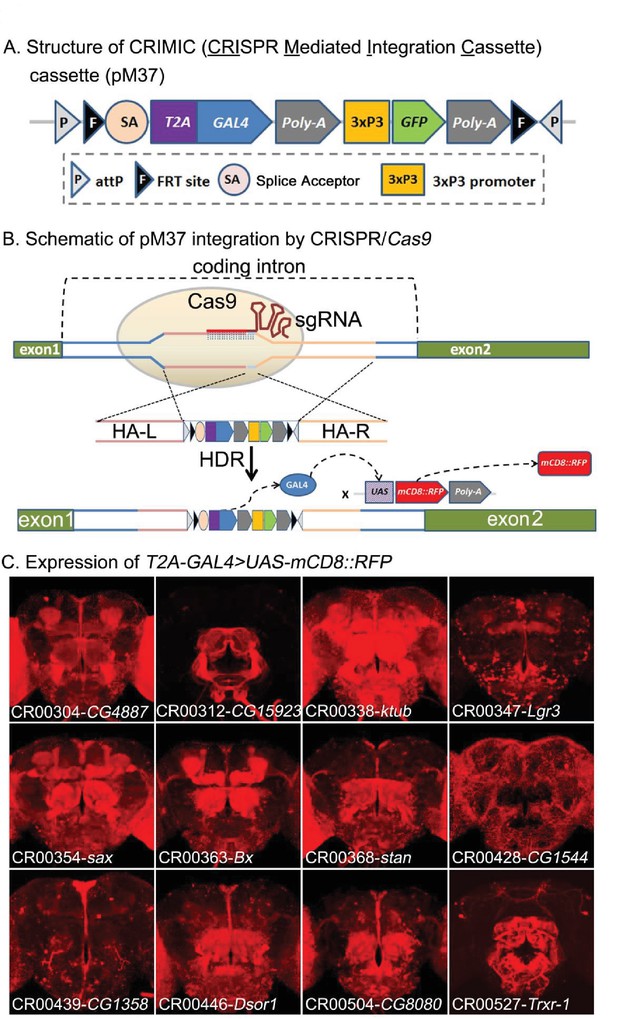
CRIMIC: T2A-GAL4 integration using CRISPR and expression patterns of tagged genes.
(A) Structure of the CRIMIC pM37 cassette. (B) Schematic of the CRIMIC insertion strategy through two 1 kb homology arms by HDR (homology directed repair) based on CRISPR/Cas9 technology. (C) Expression patterns observed in adult fly brains of T2A-GAL4 > UAS-mCD8::RFP. mCD8::RFP (red). Scale bar: 50 μm.
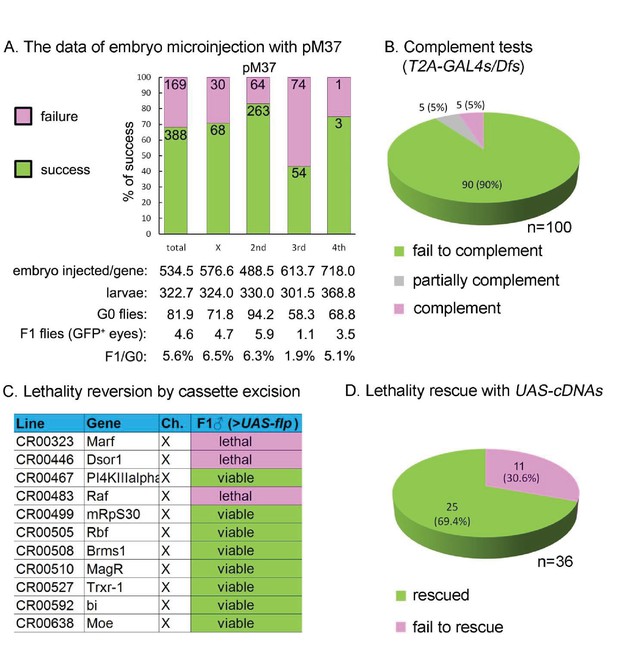
Summary of CRIMIC T2-GAL4 integration efficiency and genetic properties of T2A-GAL4 insertions (A) microinjection success rates for pM37.
(B) Complementation test: 90% of the T2A-GAL4 containing chromosomes fail to complement the corresponding Dfs; 5% produced less than 1/3 of the expected progeny; and 5% fully complemented the Dfs. For details see Supplemental Information 2. (C) T2A-GAL4 cassette excision. The lethality associated with 8 out 11 insertions is reverted in the presence of UAS-FLP. (D) Rescue of the lethality of the T2A-GAL4 cassette insertions with UAS-cDNA.
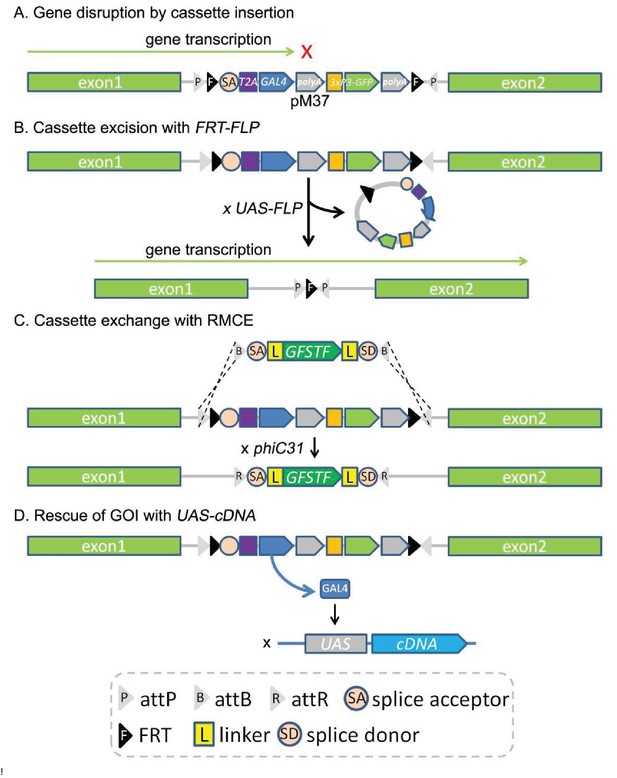
Applications of the CRIMIC technology.
(A) The CRIMIC pM37 cassette contains polyA 3’ of GAL4 which will arrest transcription and typically generate a severe loss-of-function allele. (B) The lethality caused by SA-T2A-GAL4-polyA cassette insertion can often be reverted by cassette excision using the FRT sites at the ends of the cassette and FLP. (C) The inserted SA-T2A-GAL4-polyA cassette can be replaced with other DNA fragments containing two attB sites and phiC31 integrase by RMCE such as GFSTF. (D) The SA-T2A-GAL4-polyA insertion produces GAL4 that can be used to rescue the phenotype (lethality caused by the cassette insertion) with UAS-cDNA.
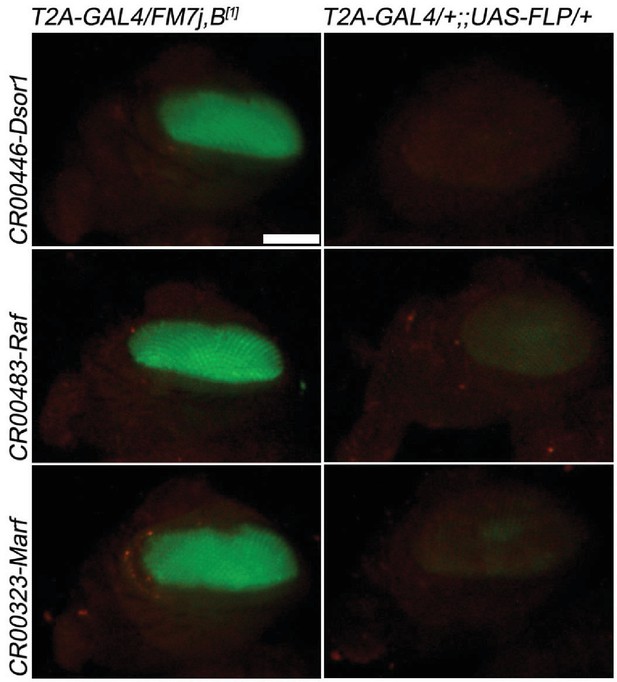
T2A-GAL4 cassette excision upon FRT-FLP.
CRIMIC T2A-GAL4 cassette, pM37 contains two FRT sites. The cassettes in three genes (Dsor1, Raf, Marf) failed to revert the lethality when exposed to UAS-FLP. However, cassette excision is still effective based on the loss of the 3XP3 Eye-GFP in Dsor1; however, weak GFP expression remains in Raf and Marf. Left panel: CRIMIC T2A-GAL4s; Right panel: CRIMIC T2A-GAL4s > UAS FLP. Scale bar: 0.2 mm.
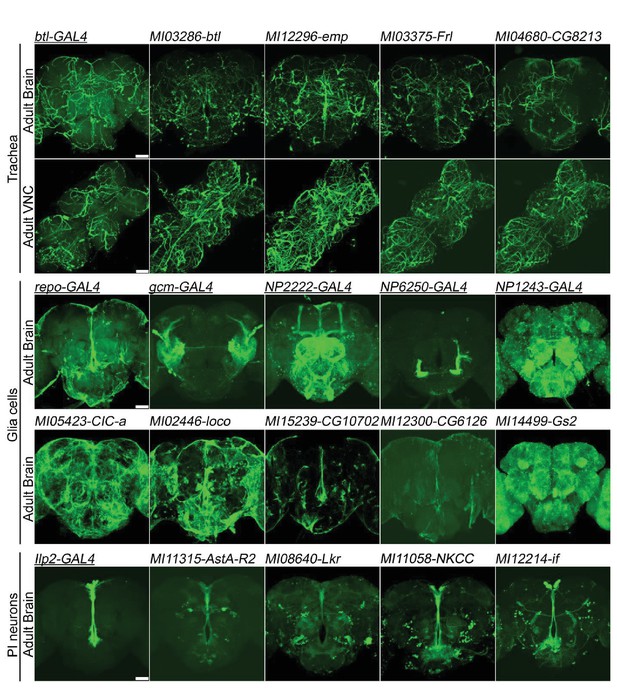
Genes expressed in (A) trachea, (B) glial cells, and (C) Pars Intercerebralis Neurons based on T2-GAL4 insertions.
The GAL4s (underlined) are existing P-element enhancer traps expressing GAL4 in specific cell populations and serve as controls. mCD8::GFP: green. Scale bar: 50 μm.
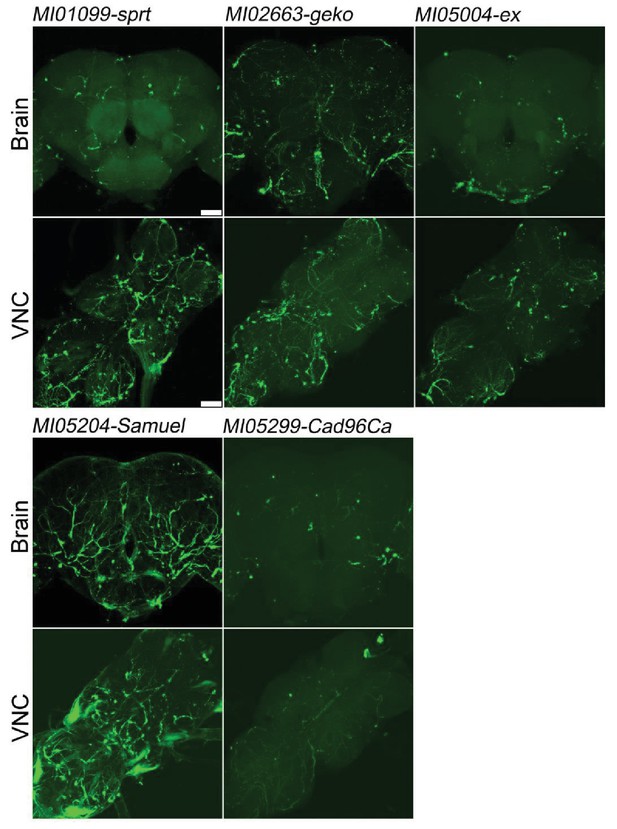
Genes specifically expressed in trachea.
Five genes previously not reported to be expressed in trachea (sprt, geko, ex, Samuel, Cad96Ca). Expression in trachea in adult brains and VNC. MiMIC T2A-GAL4s were crossed with UAS-mCD8::GFP (green). Scale bar: 50 μm.
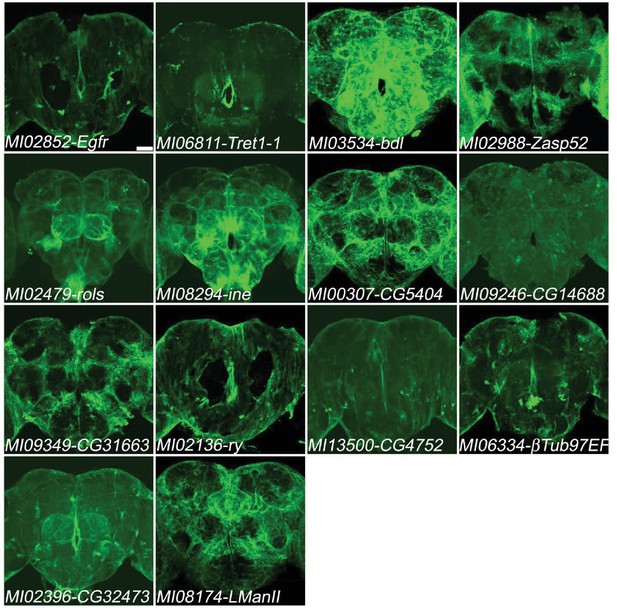
Genes expressed in glia.
Two genes (Egfr, Tret1-1) previously documented to be expressed in glia and twelve genes not previously documented to be expressed in glia (bdl, Zasp52, rols, ine, CG5404, CG14688, CG31663, ry, CG4752,βTub97EF, CG32473, LManII) in adult brains. MiMIC T2A- GAL4s were crossed with UAS-mCD8::GFP (green). Note that the damages were caused by dissections in MI02852-Egfr and MI02136-ry. Scale bar: 50 μm.
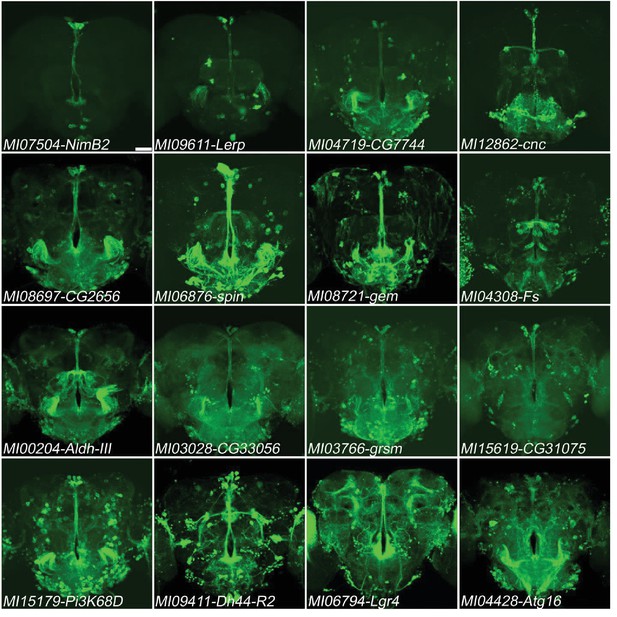
Genes expressed in PI neurons.
16 genes (NimB2, Lerp, CG7744, cnc, CG2656, spin, gem, Fs, Aldh-III, CG33056, grsm, CG31075, Pi3K68D, Dh44-R2, Lgr4, Atg16) exhibit relatively specific expression in PI neurons in adult brains. MiMIC T2A-GAL4s were crossed with UAS-mCD8::GFP (green). Scale bar: 50 μm.
Tables
Rescue of the lethality of T2A-GAL4s insertions/Dfs with aUAS-cDNA and genomic duplications with P[acman] clones.
*1:(Luo et al., 2017)*2:(Chao et al., 2017)*3:(Yoon et al., 2017)*4:(Sandoval et al., 2014). Note that a failure to rescue lethality does not mean that it cannot partially rescue other scorable phenotypes.
| Flies for rescue | ||||||
|---|---|---|---|---|---|---|
| Line | Gene | Chr. | Protein isoforms | Flies for complementation test | Fly cDNA | Genomic DNA |
| MI01374-TG4.0 | sbr | X | 1 | NA | no tag | Dp(1;3)DC508 |
| MI02836-TG4.0 | cac*1 | X | 8 | NA | EGFP | Dp(1;3)DC131 |
| MI07818-TG4.0 | acj6 | X | 13 | NA | 3xHA | Dp(1;3)DC192 |
| MI08675-TG4.1 | arm | X | 2 | NA | 3xHA | Dp(1;3)DC034 |
| MI10323-TG4.1 | flw | X | 2 | NA | 1xHA | Dp(1;3)DC224 |
| MI12214-TG4.2 | if | X | 2 | NA | no tag | Dp(1;3)DC319 |
| MI00783-TG4.0 | stj | 2 | 3 | Df(2R)Exel7128/CyO | 3xHA | NA |
| MI02963-TG4.0 | CAP | 2 | 20 | Df(2R)BSC281/CyO | no tag | NA |
| MI03306-TG4.1 | kuz | 2 | 4 | Df(2L)BSC147/CyO | no tag | NA |
| MI03597-TG4.1 | mol | 2 | 2 | Df(2R)Exel6066/CyO | 3xHA | NA |
| MI04800-TG4.1 | lola | 2 | 20 | Df(2R)ED2076/SM6a | 3xHA | NA |
| MI06876-TG4.1 | spin | 2 | 3 | Df(2R)Jp8, w[+]/CyO | myc-EGFP | NA |
| MI09180-TG4.1 | Bsg | 2 | 2 | Df(2L)ED548/SM6a | 3xHA | NA |
| MI09585-TG4.1 | Lpt | 2 | 2 | Df(2R)BSC610/SM6a | 1xHA | NA |
| MI13162-TG4.0 | Rho1 | 2 | 1 | Df(2R)ED2457/SM6a | 3xHA | NA |
| MI13708-TG4.0 | Cka | 2 | 4 | P{ry[+t7.2]=PZ}Cka[05836] cn[1]/CyO | EGFP | NA |
| MI15480-TG4.2 | kn*2 | 2 | 5 | Df(2R)BSC429/CyO | 3xHA | NA |
| MI02220-TG4.1 | dally | 3 | 1 | Df(3L)ED4413/TM6C, cu[1] Sb[1] | no tag | NA |
| MI04910-TG4.1 | ftz-f1 | 3 | 3 | Df(3L)BSC844/TM6C, Sb[1] cu[1] | 3xHA | NA |
| MI06026-TG4.1 | Nc73EF*3 | 3 | 3 | Df(3L)ED4685/TM6C,cu[1] Sb[1] | Flag | Dp(1;3)DC245 |
| MI07056-TG4.0 | Atg1 | 3 | 2 | Df(3L)BSC613/TM6C, cu[1] Sb[1] | no tag | NA |
| MI08143-TG4.0 | Sod1 | 3 | 2 | Df(3L)BSC817/TM6C, Sb[1] cu[1] | no tag | NA |
| MI05068-TG4.0 | kdn | X | 2 | NA | NA | Dp(1;3)DC154 |
| Line | Gene | Chr. | Transcripts | Df | Fly cDNA | Genomic DNA |
| CR00323 | Marf | X | 2 | NA | 1xHA*4 | Dp(1;3)DC155 |
| CR00446 | Dsor1 | X | 2 | NA | 3xHA | Dp(1;3)DC205 |
| CR00483 | Raf | X | 1 | NA | no tag | Dp(1;3)DC404 |
| CR00505 | Rbf | X | 1 | NA | 3xHA | Dp(1;3)DC012 |
| CR00638 | Moe | X | 7 | NA | myc | Dp(1;3)DC199 |
| CR00354 | sax | 2 | 3 | Df(2R)BSC265/CyO | 3xHA | NA |
| CR00465 | Dap160 | 2 | 6 | Df(2L)BSC302/CyO | no tag | NA |
| CR00466 | Eps-15 | 2 | 4 | Df(2R)BSC606/SM6a | no tag | NA |
| CR00494 | l(2)gd1 | 2 | 2 | Df(2L)Exel6027/CyO | 1xHA | NA |
| CR00521 | Npc1a | 2 | 2 | Df(2L)BSC143/CyO | YFP | NA |
| CR00559 | Sod2 | 2 | 1 | Df(2R)Exel7145/CyO | no tag | NA |
| CR00587 | Hr38 | 2 | 2 | Df(3R)BSC510/TM6C, Sb[1] cu[1] | 3xHA | NA |
| CR00762 | Wee1 | 2 | 1 | Df(2L)BSC108/CyO | no tag | NA |
| CR00452 | sr | 3 | 4 | Df(3R)BSC510/TM6C, Sb[1] cu[1] | no tag | NA |
-
Blue: fail to complement
Gray: partially complement
-
Green: rescued
Pink: fail to rescue
-
Orange: rescue phenotype but not lethality
Additional files
-
Supplementary file 1
CRISPR crosses.
- https://doi.org/10.7554/eLife.35574.017
-
Supplementary file 2
Information of MiMIC/CRIMIC lines, fly stocks, complementation results and resource.
- https://doi.org/10.7554/eLife.35574.018
-
Supplementary file 3
DNA sequences of CRIMIC donor vectors.
- https://doi.org/10.7554/eLife.35574.019
-
Transparent reporting form
- https://doi.org/10.7554/eLife.35574.020





