The origin of the odorant receptor gene family in insects
Figures
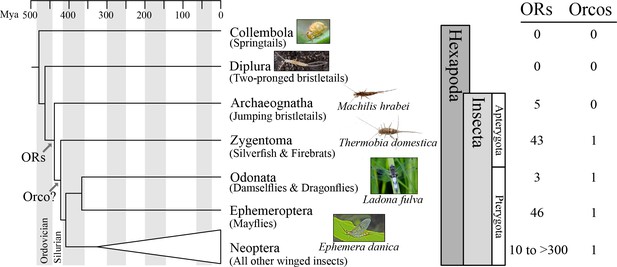
Origin of the insect odorant receptor gene family.
The number of ORs and OR co-receptors (Orcos) for all species of the insect and other hexapod orders analyzed was mapped on the hexapod phylogeny sensu (Misof et al., 2014). ORs are present in all insects but absent from non-insect hexapod genomes, and thus likely represent an evolutionary novelty for the Class Insecta. Orco is present in all but Archaeognatha, an ancestrally wingless (apterygote) insect order. This suggests two scenarios including either the loss of Orco in Archaeognatha or an Orco origin following the evolution of ORs (as indicated). The genomes of all neopteran insects analyzed to date encode ORs, ranging from 10 ORs in head lice (Kirkness et al., 2010) to more than 300 ORs in ants (Smith et al., 2011a, 2011b).
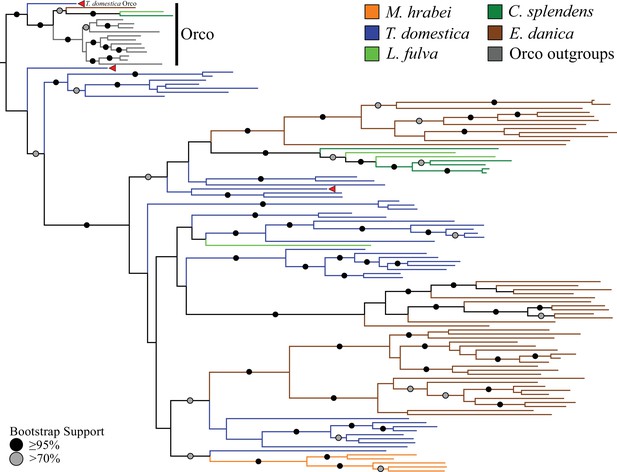
Odorant receptor (OR) gene family phylogeny including representatives of all apterygote and paleopteran insect orders.
The Maximum Likelihood tree demonstrates monophyly of the single-copy insect Orco with high bootstrap support. The M. hrabei genome lacks Orco but encodes five ORs clustering in a single highly-supported clade. T. domestica has a fully developed functional OR/Orco system. The red arrowheads indicate the locations of the three T. domestica ORs identified by Missbach et al. (2014), including the gene identified as T. domestica Orco in this study (formerly TdomOrco2).
Additional files
-
Supplementary file 1
Table S1 Transposable element repeat class analysis of the Thermobia domestica genome assembly. Table S2 Details of the Thermobia domestica OR family genes and proteins. Columns are: Gene – the gene and protein name we are assigning (suffixes, which are not part of the name but indicate features of the gene model, are C – C-terminus missing, F – assembly was repaired, J – gene model spans scaffolds, * - one or more join across scaffolds made on the basis of comparison with an ortholog in Ctenolepisma longicaudata or a close intact relative in Thermobia); Scaffold – the v1 genome assembly scaffold ID; Coordinates – the nucleotide range from the first position of the start codon to the last position of the stop codon in the contig/scaffold; Strand –+ is forward and - is reverse; RNA – number of independent pairs of reads from Missbach et al. (2014) and 1Kite (Misof et al., 2014); Introns – phases of introns (bold indicates those supported by Missbach et al. cDNAs, or their raw RNAseq reads, or those from 1Kite); % - percent identity for most apparent 1–1 orthologs with Ctenolepisma longicaudata; AAs – number of encoded amino acids in the protein; Comments – comments on the gene model. Note that Orco is Orco2, Or1 is Orco1, and Or9 is Orco3 of Missbach et al. (2014). Table S3 Details of the MhraOr family genes and proteins. Columns are: Gene – the gene and protein name we are assigning (suffixes, which are not part of the name but indicate features of the gene model, are F – assembly was repaired, J – gene model spans scaffolds, * - one or more joins across scaffolds is based only on sequence similarity to the other proteins); Scaffold – the v1 genome assembly scaffold ID from i5k; Coordinates – the nucleotide range from the first position of the start codon to the last position of the stop codon; Strand –+is forward and - is reverse; RNA – number of independent pairs of reads from 1Kite and the i5k pilot project (single reads from Missbach et al. (2014) for the related species Lepismachilis y-signata are shown in parentheses); Introns – phases of introns (bold indicates those supported by RNAseq reads); AAs – number of encoded amino acids in the protein; Comments – comments on the gene model. FASTA format proteins for the newly described ORs: Suffixes, which are not part of the gene/protein name but indicate features of the gene model, are C – C-terminus missing, F – assembly was repaired, I – internal regions missing, J – gene model spans scaffolds, P – pseudogene. All proteins of the newly described ORs and the alignment used to reconstruct the gene tree are available on Dryad.
- https://doi.org/10.7554/eLife.38340.004
-
Supplementary file 2
kmer frequency spectrum of Thermobia domestica sequencing reads.
A high heterozygous peak with a maximum at k = 17 in comparison to the homozygous peak around k = 37 indicates high heterozygosity in the genomic data. High heterozygosity is a known culprit to difficult genome assembly but cannot be avoided in most non-model organisms, which often cannot be used to produce inbred lines.
- https://doi.org/10.7554/eLife.38340.005
-
Supplementary file 3
Odorant receptor gene family phylogeny of all apterygote and paleopteran insect orders.
The Maximum Likelihood phylogeny shows relationships between the ORs and Orcos detected in M. hrabei (orange), T. domestica (blue), L. fulva (bright green), C. splendens (dark green; Ioannidis et al., 2017), and E. danica (brown).
- https://doi.org/10.7554/eLife.38340.006
-
Transparent reporting form
- https://doi.org/10.7554/eLife.38340.007





