Control of feeding by Piezo-mediated gut mechanosensation in Drosophila
Figures
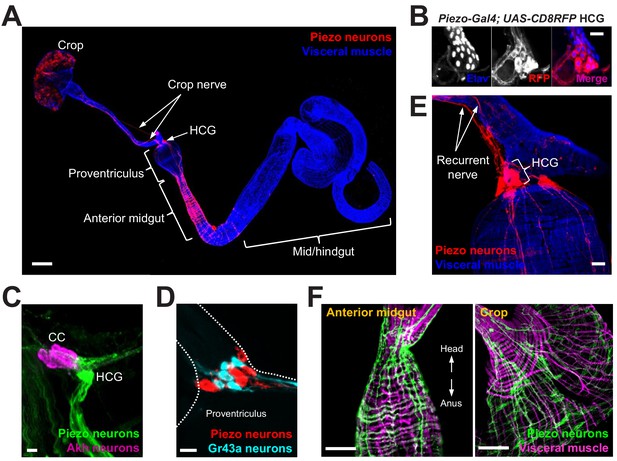
Piezo neurons innervate the gastrointestinal tract.
(A) Wholemount image of the digestive tract from a Piezo-Gal4 (59266); UAS-DenMark fly visualized with immunofluorescence for DenMark (red, anti-Red Fluorescent Protein or RFP) and a fluorescent Phalloidin conjugate (blue) to label visceral muscle. HCG: hypocerebral ganglion, scale bar 100 μm. (B) Immunofluorescence for RFP (red) and Elav (blue) in the HCG from a Piezo-Gal4; UAS-CD8RFP fly, scale bar 10 μm. (C) Immunofluorescence for GFP (green) and Akh (magenta) in the corpora cardiaca (CC) and HCG from a Piezo-Gal4; UAS-CD8GFP fly, scale bar 10 μm. (D) Native GFP and RFP fluorescence from the HCG of a Piezo-Gal4; UAS-CD8RFP; Gr43a-LexA; LexAop-CD8GFP fly, scale bar 10 μm. (E) Image of the recurrent nerve (arrows) labeled by native RFP fluorescence in a Piezo-Gal4; UAS-CD8RFP fly and a fluorescent Phalloidin conjugate (blue), scale bar 10 μm. (F) The anterior midgut (left) and crop (right) of a Piezo-Gal4; UAS-DenMark fly visualized by immunofluorescence for DenMark (green) and a fluorescent Phalloidin conjugate (magenta), scale bar 50 μm. See Figure 1—figure supplement 1 and source data.
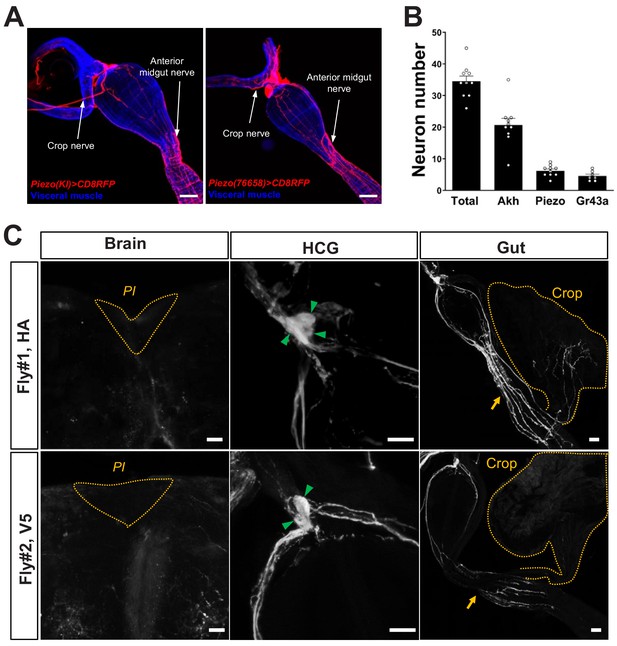
Innervation of the gastrointestinal tract by Piezo neurons.
(A) Wholemount image of the digestive tract from two additional Piezo-Gal4; UAS-CD8RFP fly lines visualized with native RFP fluorescence (red) and a fluorescent Phalloidin conjugate (blue) to label visceral muscle. HCG: hypocerebral ganglion, scale bar 50 μm. (B) The average number of neurons in the hypocerebral ganglion and corpora cardiaca labeled per fly by immunofluorescence of Elav (total), Akh (AKH), and RFP (Piezo) in Piezo-Gal4; UAS-CD8RFP flies, and native GFP fluorescence (Gr43a) in Gr43a-LexA; LexAop-CD8GFP flies. n (left to right): 10, 10, 10, and 7 flies, mean ± SEM. (C) Sparse labeling of Piezo neurons was achieved in Piezo-Gal4 (59266) flies crossed with MultiColor FlpOut (MCFO) flies (Hs-Flp; UAS-MCFO flies) by brief warming (37°C, 15 min/day, 3 days). In example flies shown, wholemount immunostaining for HA (top) and V5 (bottom) epitopes is depicted in the pars intercerebralis (PI, dotted orange lines) of the brain, the hypocerebral ganglion (HCG, green arrows: labeled soma), and the gut (yellow arrows: location of the anterior midgut; yellow dotted lines: crop), scale bars 20 μm.
-
Figure 1—figure supplement 1—source data 1
Numerical data to support the graph in Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/63049/elife-63049-fig1-figsupp1-data1-v2.xlsx
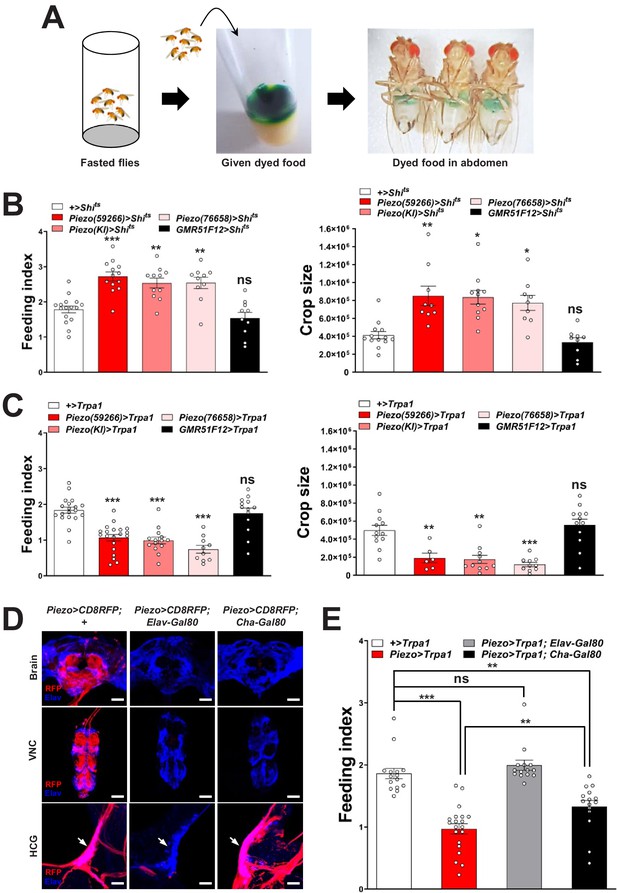
Piezo neurons control feeding behavior.
(A) Depiction of the colorimetric feeding assay. (B) Fasted flies with Shibire alleles indicated were given brief access (30 min) to dye-labeled food at 32°C, and feeding indices and crop sizes were calculated. n (left to right) (feeding index): 16, 11, 13, 10, and 10 trials involving 12 flies per trial. n (crop size): 13, 9, 11, 9, and 9 flies, mean ± SEM, ***p<0.0005, **p<0.005, *p<0.05, ns: not significant by ANOVA Dunnett’s multiple comparison test. (C) Fasted flies with Trpa1 alleles indicated were given brief access (30 min) to dye-labeled food at 30°C, and feeding indices and crop sizes were calculated. n (left to right) (feeding index): 19, 20, 14, 10, and 13 trials involving 12 flies per trial. n (crop size): 12, 6, 11, 10, and 12 flies, mean ± SEM, ***p<0.0005, **p<0.005, ns: not significant by ANOVA Dunnett’s multiple comparison test. (D) Native RFP fluorescence in brain (top), ventral nerve cord (VNC, middle), and hypocerebral ganglion (HCG, bottom) of Piezo-Gal459266; UAS-CD8RFP flies with Gal80 alleles indicated, scale bar 100 μm (brain, VNC), 20 μm (HCG). (E) Fasted flies with Trpa1 alleles indicated were given brief access (30 min) to dye-labeled food at 30°C, and feeding indices were calculated. n (left to right): 15, 20, 14, and 15 trials involving 12 flies per trial, mean ± SEM, ***p<0.0005, **p<0.005, ns: not significant by ANOVA Dunnett’s multiple comparison test. See Figure 2—figure supplements 1–3 and source data.
-
Figure 2—source data 1
Numerical data to support the graphs in Figure 2.
- https://cdn.elifesciences.org/articles/63049/elife-63049-fig2-data1-v2.xlsx
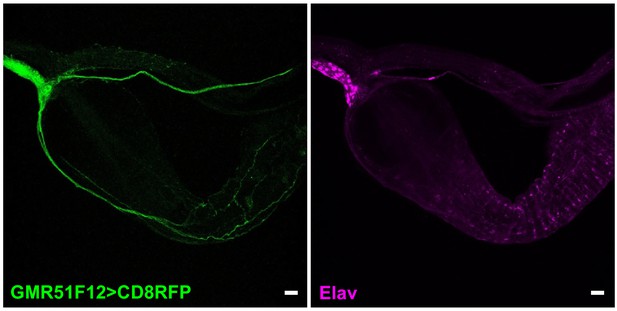
Visualizing gut innervation by neurons labeled in GMR51F12-Gal4 flies.
Wholemount immunostaining for RFP (left) and Elav (right) in gut of GMR51F12-Gal4; UAS-CD8RFP flies, scale bars 20 μm.
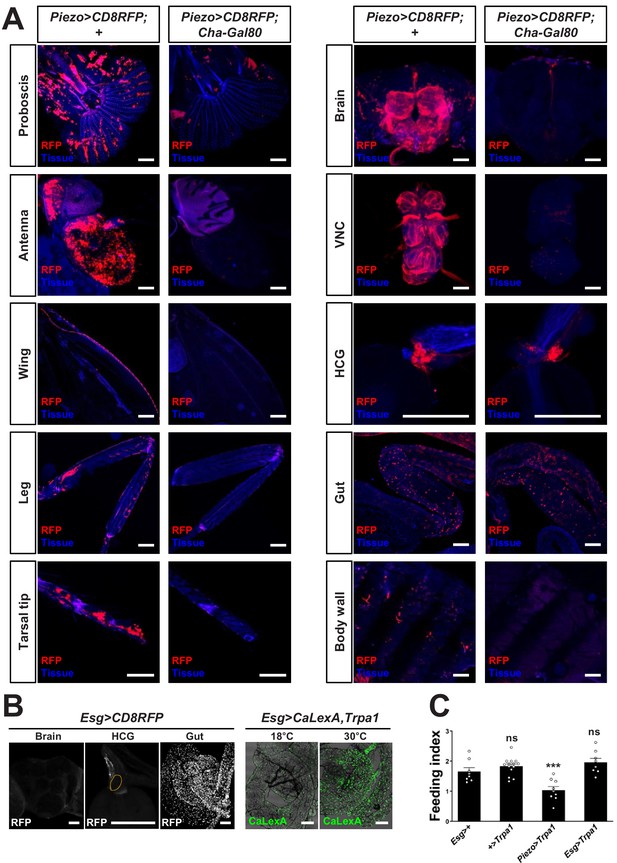
Visualizing and manipulating subtypes of Piezo neurons.
(A) Native RFP fluorescence in tissues indicated from Piezo-Gal4 (59266); UAS-CD8RFP flies with or without Cha-Gal80, scale bars: 100 μm. (B) Wholemount images of native RFP fluorescence in the brain, proventriculus (orange outline: HCG), and midgut of Esg-Gal4; UAS-CD8RFP flies (left), and wholemount images of native GFP fluorescence in the midgut of Esg-Gal4, UAS-CaLexA, UAS-Trpa1 flies at 18°C and 30°C (right). Scale bars: 100 µm. (C) Fasted flies with alleles indicated were given brief access (30 min) to dye-labeled food at 30°C, and feeding indices were calculated. n (left to right): 8, 14, 9, and 8 trials involving 12 flies per trial, mean ± SEM, ***p<0.0005, ns: not significant by ANOVA Dunnett’s multiple comparison test.
-
Figure 2—figure supplement 2—source data 1
Numerical data to support the graph in Figure 2—figure supplement 2.
- https://cdn.elifesciences.org/articles/63049/elife-63049-fig2-figsupp2-data1-v2.xlsx
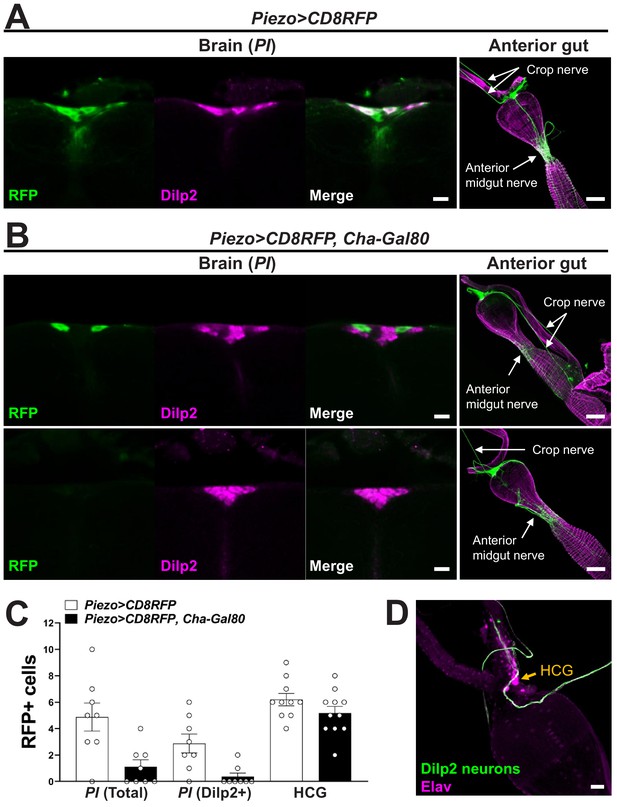
Analyzing central and peripheral cell types labeled in various genetic models.
(A) Brain: wholemount immunostaining for RFP (green) and Dilp2 (magenta) in the pars intercerebralis (PI) region of Piezo-Gal4; UAS-CD8RFP flies, scale bar 10 µm; anterior gut: wholemount immunostaining for RFP (green) and Phalloidin (magenta) in the gut of Piezo-Gal4; UAS-CD8RFP flies, scale bar 50 µm. (B) Brain: wholemount immunostaining for RFP (green) and Dilp2 (magenta) in the PI region of Piezo-Gal4; UAS-CD8RFP; Cha-Gal80 flies, scale bar 10 µm; anterior gut: wholemount immunostaining for RFP (green) and Phalloidin (magenta) in the gut of Piezo-Gal4; UAS-CD8RFP; Cha-Gal80 flies, scale bar 50 µm. (C) The number of RFP-labeled cells in the PI, total or co-labeled with Dilp2, or in the hypocerebral ganglion (HCG). n (left to right): 8, 8, 8, 8, 10, and 11 flies, mean ± SEM. (D) Wholemount immunostaining for GFP (green) and Elav (magenta) in the hypocerebral ganglion (arrow) of Dilp2-Gal4; UAS-CD8GFP flies, scale bar 20 µm.
-
Figure 2—figure supplement 3—source data 1
Numerical data to support the graph in Figure 2—figure supplement 3.
- https://cdn.elifesciences.org/articles/63049/elife-63049-fig2-figsupp3-data1-v2.xlsx
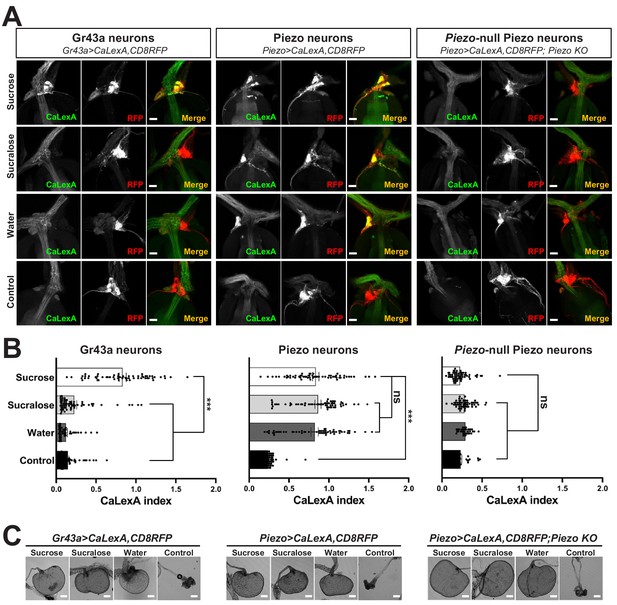
Piezo mediates enteric neuron responses to crop-distending stimuli.
(A) Flies of genotypes indicated were provided solutions of (1) sucrose, (2) sucralose, (3) water alone after a period of water deprivation (water), or (4) water alone ad libitum for 24 hr (control). Representative images of native CaLexA-induced GFP reporter (green) and CD8RFP (red) fluorescence visualized in enteric Gr43a neurons (left), Piezo neurons (middle), or Piezo neurons lacking Piezo (right), scale bar 10 μm. (B) Quantification of CaLexA-induced GFP fluorescence in individual RFP-expressing neurons from flies in (A). n (from top to bottom): 59, 64, 43, and 67 Gr43a neurons from 13, 14, 9, and 15 flies; 61, 61, 59, and 66 Piezo neurons from 11, 11, 10, and 12 flies; 60, 60, 33, and 37 Piezo-null Piezo neurons from 11, 11, 5, and 6 flies, mean ± SEM, ***p<0.0001, ns: not significant by ANOVA Dunnett’s multiple comparison test. (C) Visualization of the crop from flies given stimuli indicated after 24 hr (sucrose, sucralose, control) or 15 min (water), scale bar 100 μm. See Figure 3—figure supplement 1 and source data.
-
Figure 3—source data 1
Numerical data to support the graph in Figure 3.
- https://cdn.elifesciences.org/articles/63049/elife-63049-fig3-data1-v2.xlsx
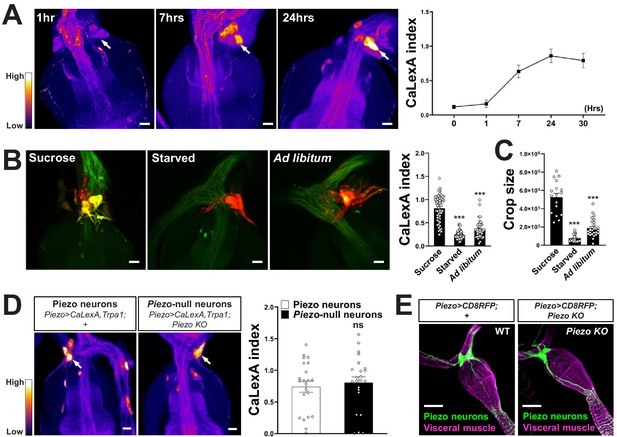
Responses and innervation patterns of Piezo neurons in wild-type and Piezo knockout flies.
(A) Piezo-Gal4; UAS-CaLexA; UAS-Trpa1 flies were placed at 30°C for indicated time periods with ad libitum food. Representative pseudocolor images (left) and quantification (right) of native CaLexA-induced GFP reporter fluorescence in the hypocerebral ganglion. GFP fluorescence was visually transformed to a color map indicating fluorescence intensity. n (left to right): 10, 10, 18, 11, and 12 flies, mean ± SEM, scale bar 10 μm. (B) Piezo-Gal4; UAS-CaLexA; UAS-CD8RFP flies were provided for 24 hr with sucrose, water alone (starved), or regular food ad libitum (ad libitum). Representative images (left) of native CaLexA-induced GFP reporter (green) and CD8RFP (red) fluorescence, and quantification (right) of CaLexA-induced GFP fluorescence in individual RFP-expressing neurons. n (left to right): 54, 82, and 44 neurons from 9, 16, and 8 flies, mean ± SEM, ***p<0.0001, ns: not significant by ANOVA Dunnett’s multiple comparison test, scale bar 10 μm. (C) Crop sizes from wild-type flies fed as indicated. n (left to right): 17, 19, and 28 flies, mean ± SEM, ***p<0.0001 by ANOVA Dunnett’s multiple comparison test. (D) Piezo-Gal4; UAS-CaLexA; UAS-Trpa1 (Piezo neurons) and Piezo-Gal4; UAS-CaLexA; UAS-Trpa1; Piezo knockout (Piezo-null neurons) flies were placed at 30°C with ad libitum food for 24 hr. Representative pseudocolor images (left) and quantification (right) of native CaLexA-induced GFP reporter fluorescence in the hypocerebral ganglion. GFP fluorescence was visually transformed to a color map indicating fluorescence intensity. n: 22 (left) or 23 (right) flies, mean ± SEM, ns: not significant by unpaired t-test, scale bar 10 μm. (E) Wholemount images of the digestive tract from Piezo-Gal4; UAS-CD8RFP and Piezo-Gal4; UAS-CD8RFP; Piezo knockout flies visualized with immunofluorescence for RFP (green, anti-RFP) and a fluorescent Phalloidin conjugate (magenta) to label visceral muscle, scale bar 50 μm.
-
Figure 3—figure supplement 1—source data 1
Numerical data to support the graph in Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/63049/elife-63049-fig3-figsupp1-data1-v2.xlsx
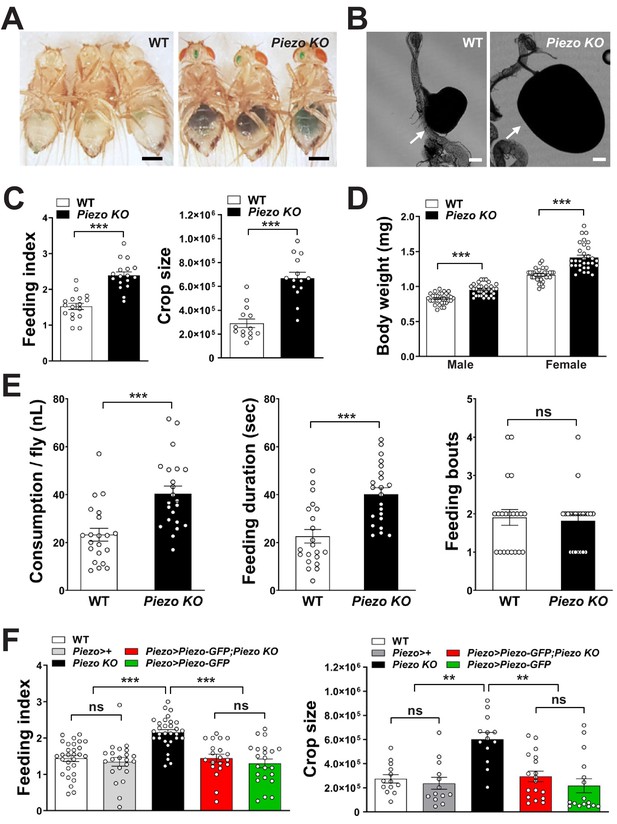
Piezo knockout alters fly feeding behavior.
(A) Fasted wild-type (WT) and Piezo knockout (KO) female flies were given brief access (30 min) to dye-colored food and imaged, scale bar 0.5 mm. (B) Representative images of the crop (arrow) in WT and Piezo KO flies, scale bar 100 μm, (C) Calculated feeding indices (left) and crop sizes (right) from flies in (A). n (feeding index: 17 trials involving 204 flies), n (crop size): 14 flies, mean ± SEM, ***p<0.0001 by unpaired t-test. (D) Body weights of WT and Piezo KO flies fed regular food ad libitum. n (left to right): 32, 34, 33, and 31 trials involving three flies per trial, mean ± SEM, ***p<0.0001 by unpaired t-test. (E) Feeding parameters of fasted WT and Piezo KO male flies were analyzed using the EXPRESSO assay for 30 min after food introduction to determine overall food consumption, feeding duration per bout, and the number of bouts. n: 21 (WT), 22 (PIEZO KO) flies, mean ± SEM, ***p<0.0005, ns: not significant by unpaired t-test. (F) Calculated feeding indices (left) and crop sizes (right) from Piezo rescue and control flies indicated. n (left to right) (feeding index): 29, 22, 30, 22, and 13 trials involving 12 flies per trial. n (crop size): 13, 13, 13, 18, and 16 flies, mean ± SEM, ***p<0.0005, **p<0.005 by ANOVA Dunnett’s multiple comparison test, ns: not significant by unpaired t test. See Figure 4—figure supplements 1 and 2 and source data.
-
Figure 4—source data 1
Numerical data to support the graph in Figure 4.
- https://cdn.elifesciences.org/articles/63049/elife-63049-fig4-data1-v2.xlsx
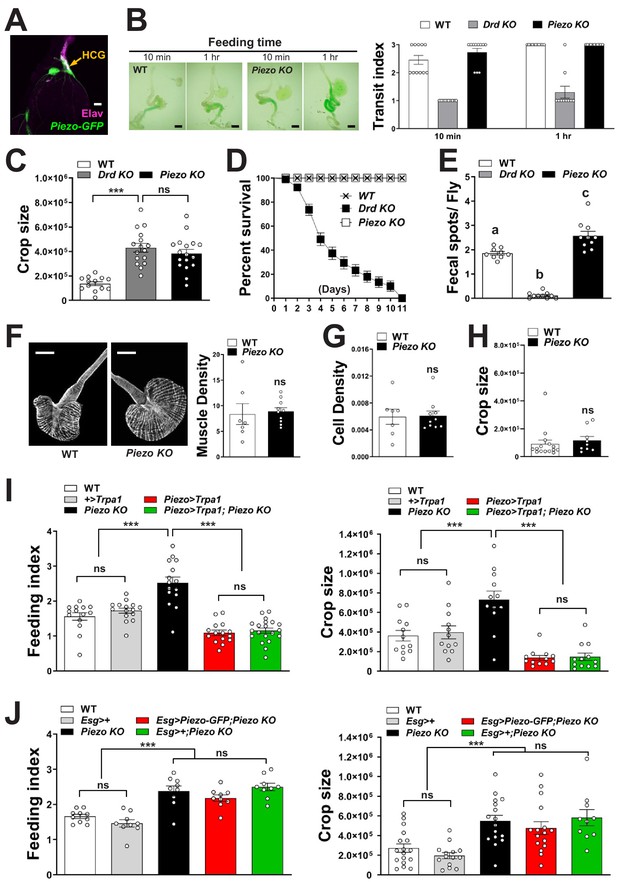
Physiological characterization of Piezo knockout flies.
(A) Immunofluorescence for GFP (green) and Elav (magenta) in the hypocerebral ganglion (HCG) of Piezo rescue flies (Piezo knockout; Piezo-Gal4 [59266]; UAS-Piezo-GFP), scale bar 20 μm. (B) Visualizing (left) and quantifying (right) intestinal transit of dye-colored food in wild-type (WT), Drop-dead knockout (Drd KO), and Piezo knockout (Piezo KO) flies. n (left to right): 11, 11, 10, 11, 11, and 10 flies, mean ± SEM, scale bar 200 μm, differences between WT and Piezo KO were not significant by either ANOVA Dunnett’s multiple comparison test or by Fisher's exact test. Measurements of (C) crop size, (D) survival, and (E) fecal spot deposition in WT, Piezo KO, and Drop-dead KO flies. n (left to right) for (C): 13, 16, and 17 flies; for (D): 8, 7, and 7 trials involving 12 flies per trial for WT, Drd KO, and Piezo KO involving 84–96 flies; for (E): 9, 10, and 10 trials involving 10 flies per trial, mean ± SEM, ***p<0.0001; ns: not significant by ANOVA Dunnett’s multiple comparison test; statistical comparison by ANOVA Dunnett’s multiple comparison test of (a)–(c): p=0.0006, of (a, b): p<0.0001. (F) Visualization (left) and quantification (right) of muscle fiber density by a fluorescent Phalloidin conjugate, n: 7 (left) and 10 (right) flies, mean ± SEM, scale bar 100 μm. (G) Quantification of crop cell density based on nuclear staining (TO-PRO-3), n: 7 (left) and 10 (right) flies, mean ± SEM. (H) Measurement of crop size in fasted flies, n: 15 (left) and 9 (right) flies, mean ± SEM. (I) Fasted flies of genotypes indicated were given brief access (30 min) to dye-labeled food at 30°C, and feeding indices and crop sizes were calculated. n (left to right) (feeding index): 14, 15, 15, 15, and 20 trials involving 12 flies per trial. n (crop size): 12 flies, mean ± SEM, ***p<0.0005, ns: not significant by ANOVA Dunnett’s multiple comparison test or unpaired t-test. (J) Fasted flies of genotypes indicated were given brief access (30 min) to dye-labeled food, and feeding indices and crop sizes were calculated. n (left to right) (feeding index): 10, 9, 9, 9, 9, and 9 trials involving 12 flies per trial. n (crop size): 17, 15, 14, 16, 16, and 10 flies, mean ± SEM, ***p<0.0005, ns: not significant by ANOVA Dunnett’s multiple comparison test.
-
Figure 4—figure supplement 1—source data 1
Numerical data to support the graph in Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/63049/elife-63049-fig4-figsupp1-data1-v2.xlsx
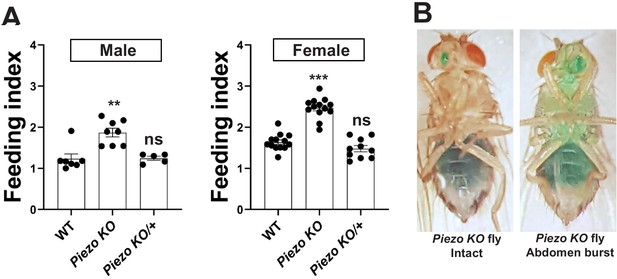
Feeding characteristics of Piezo knockout flies.
(A) Feeding indices of male and female wild-type (WT) and Piezo knockout homozygote (Piezo KO) and heterozygote (Piezo KO/+) flies. Flies were fasted, given brief access (30 min) to dye-colored food, and scored. n (left to right) for male: 7, 8, and 5 and female: 13, 13, and 10 trials involving 12 flies per trial, mean ± SEM, **p<0.0005, ***p<0.0001, ns: not significant by ANOVA Dunnett’s multiple comparison test. (B) Piezo KO flies were fasted and then fed with green dye-labeled food; images are depicted of flies with either intact abdomens (left) or bursted abdomens (right).
-
Figure 4—figure supplement 2—source data 1
Numerical data to support the graph in Figure 4—figure supplement 2.
- https://cdn.elifesciences.org/articles/63049/elife-63049-fig4-figsupp2-data1-v2.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Drosophila melanogaster) | Piezo-Gal4 | Bloomington Drosophila Stock Center | BDSC: 59266; RRID:BDSC_59266 | |
| Genetic reagent (D. melanogaster) | Piezo(KI)-Gal4 | He et al., 2018 | PMID:29414942 | |
| Genetic reagent (D. melanogaster) | Piezo(gene-trap)-Gal4 | Bloomington Drosophila Stock Center | BDSC: 76658 RRID:BDSC_76658 | |
| Genetic reagent (D. melanogaster) | Piezo KO | Bloomington Drosophila Stock Center | BDSC: 58770; RRID:BDSC_58770 | Isogenized with w1118 |
| Genetic reagent (D. melanogaster) | UAS-GFP-Piezo | Bloomington Drosophila Stock Center | BDSC: 58773; RRID:BDSC_58773 | |
| Genetic reagent (D. melanogaster) | UAS-CD8RFP | Bloomington Drosophila Stock Center | BDSC: 32218; RRID:BDSC_32218 | |
| Genetic reagent (D. melanogaster) | Hs-Flp, UAS-MCFO | Bloomington Drosophila Stock Center | BDSC: 64085; RRID:BDSC_64085 | |
| Genetic reagent (D. melanogaster) | UAS-CD8GFP | Bloomington Drosophila Stock Center | BDSC: 5137; RRID:BDSC_5137 | |
| Genetic reagent (D. melanogaster) | UAS-Trpa1 | Bloomington Drosophila Stock Center | BDSC: 26263; RRID:BDSC_26263 | |
| Genetic reagent (D. melanogaster) | UAS-CaLexA | Bloomington Drosophila Stock Center | BDSC: 66542; RRID:BDSC_66542 | |
| Genetic reagent (D. melanogaster) | Nanchung-Gal4 | Bloomington Drosophila Stock Center | BDSC: 24903; RRID:BDSC_24903 | |
| Genetic reagent (D. melanogaster) | Inactive-Gal4 | Bloomington Drosophila Stock Center | BDSC: 36360; RRID:BDSC_36360 | |
| Genetic reagent (D. melanogaster) | Painless-Gal4 | Bloomington Drosophila Stock Center | BDSC: 27894; RRID:BDSC_27894 | |
| Genetic reagent (D. melanogaster) | Tmc-Gal4 | Zhang et al., 2016 | PMID:27478019 | |
| Genetic reagent (D. melanogaster) | Gr43a-Gal4 | Miyamoto et al., 2012 | PMID:23178127 | |
| Genetic reagent (D. melanogaster) | Gr43a-LexA | Fujii et al., 2015 | PMID:25702577 | |
| Genetic reagent (D. melanogaster) | UAS-DenMark | Bloomington Drosophila Stock Center | BDSC: 33061; RRID:BDSC_33061 | |
| Genetic reagent (D. melanogaster) | UAS-DenMark | Bloomington Drosophila Stock Center | BDSC: 33062; RRID:BDSC_33062 | |
| Genetic reagent (D. melanogaster) | Trp-Gal4 | Bloomington Drosophila Stock Center | BDSC: 36359; RRID:BDSC_36359 | |
| Genetic reagent (D. melanogaster) | Nompc-Gal4 | Bloomington Drosophila Stock Center | BDSC: 36360; RRID:BDSC_36360 | |
| Genetic reagent (D. melanogaster) | Drop-dead KO | Bloomington Drosophila Stock Center | BDSC: 36360; RRID:BDSC_36360 | |
| Genetic reagent (D. melanogaster) | w1118 | Bloomington Drosophila Stock Center | BDSC: 3605; RRID:BDSC_3605 | |
| Genetic reagent (D. melanogaster) | Trpa1-Gal4 | Bloomington Drosophila Stock Center | BDSC: 36362; RRID:BDSC_36362 | |
| Genetic reagent (D. melanogaster) | Ppk-Gal4 | Bloomington Drosophila Stock Center | BDSC: 32078; RRID:BDSC_32078 | |
| Genetic reagent (D. melanogaster) | GMR51F12-Gal4 | Bloomington Drosophila Stock Center | BDSC: 58685; RRID:BDSC_58685 | |
| Genetic reagent (D. melanogaster) | Cha-Gal80 | Sakai et al., 2009 | PMID:19531155 | |
| Genetic reagent (D. melanogaster) | UAS-Shibirets | Kitamoto, 2001 | PMID:11291099 | |
| Genetic reagent (D. melanogaster) | Escargot-Gal4 | Hayashi et al., 2002 | PMID:12324948 | |
| Genetic reagent (D. melanogaster) | Elav-Gal80 | Yang et al., 2009 | PMID:19249273 | |
| Antibody | Anti-Dilp2; rabbit polyclonal | Veenstra Jan (University of Bordeaux, France) | (1:200) | |
| Antibody | Anti-GFP; chicken polyclonal | Thermo Fisher Scientific | Thermo Fisher Scientific Cat# A10262; RRID:AB_2534023 | (1:200) |
| Antibody | Anti-RFP; rabbit polyclonal | Rockland | Rockland Cat# 600-401-379; RRID:AB_2209751 | (1:200) |
| Antibody | Anti-Elav; mouse monoclonal | Developmental Studies Hydridoma Bank | DSHB Cat# Elav-9F8A9; RRID:AB_528217 | (1:200) |
| Antibody | Anti-Akh; rabbit polyclonal | Kerafast | Kerafast Cat# EGA261 | (1:200) |
| Antibody | Anti-Flag; Rat monoclonal | Novus Biologicals | Novus Cat# NBP1-06712SS; RRID:AB_1625982 | (1:200) |
| Antibody | Anti-HA; Rabbit monoclonal | Cell Signaling Technology | Cell Signaling Technology Cat# 3724S; RRID:AB_1549585 | (1:200) |
| Antibody | Anti-V5; Mouse monoclonal | Bio-Rad | Bio-Rad Cat# MCA2894D549GA RRID:AB_10845946 | (1:200) |
| Antibody | Alexa Fluor-488; Chicken polyclonal | Jackson ImmunoResearch | Jackson ImmunoResearch Cat# 703-545-155; RRID:AB_2340375 | (1:400) |
| Antibody | Alexa Fluor-488; Rabbit polyclonal | Jackson ImmunoResearch | Jackson ImmunoResearch Cat# 711-545-152; RRID:AB_2313584 | (1:400) |
| Antibody | Cy3-AffiniPure; Rabbit polyclonal | Jackson ImmunoResearch | Jackson ImmunoResearch Cat# 711-165-152; RRID:AB_2307443 | (1:400) |
| Antibody | Alexa Fluor 647; Rabbit polyclonal | Jackson ImmunoResearch | Jackson ImmunoResearch Cat# 711-605-152; RRID:AB_2492288 | (1:400) |
| Antibody | Alexa Fluor 647; Mouse polyclonal | Jackson ImmunoResearch | Jackson ImmunoResearch Cat# 715-605-150; RRID:AB_2340862 | (1:400) |
| Antibody | Alexa Fluor 488; Mouse polyclonal | Jackson ImmunoResearch | Jackson ImmunoResearch Cat# 715-545-150; RRID:AB_2340846 | (1:400) |
| Antibody | Alexa Fluor 488; Rat polyclonal | Jackson ImmunoResearch | Jackson ImmunoResearch Cat# 712-545-153; RRID:AB_2340684 | (1:400) |
| Chemical compound, drug | Normal goat serum | Jackson ImmunoResearch | Jackson ImmunoResearch Cat# 005-000-121; RRID:AB_2336990 | (5%) |
| Chemical compound, drug | Fluoromount-G | Southern Biotech | 0100-01 | |
| Chemical compound, drug | Phalloidin-FITC | Sigma | P5282-1MG | (1:400) |
| Chemical compound, drug | Phalloidin-TRITC | Sigma | P1951-1MG | (1:400) |
| Chemical compound, drug | TO-PRO-3 | ThermoFisher | T3605 | (1:400) |
| Chemical compound, drug | Green food dye | Amazon | Amazon standard identification number (ASIN): B0055AFE5G | Manufacturer: McCormick |
| Software, algorithm | Prism 8 | GraphPad | RRID:SCR_002798 | |
| Software, algorithm | Fiji | Schindelin et al., Nature Methods, 2012 | PMID:22743772 | https://imagej.net/Fiji |
| Software, algorithm | Python-based custom data analysis code used for EXPRESSO assay | Samuel C. Whitehead, 2021, PiezoPaperExpressoCode | https://github.com/scw97/PiezoPaperExpressoCode; Min, 2021; copy archived at swh:1:rev:bd8a58fa0e4f796e2ed0b72fe807862305b84b6b | |
| Other | Confocal microscope | Leica | Leica SP5 |





