The LOTUS initiative for open knowledge management in natural products research
Figures
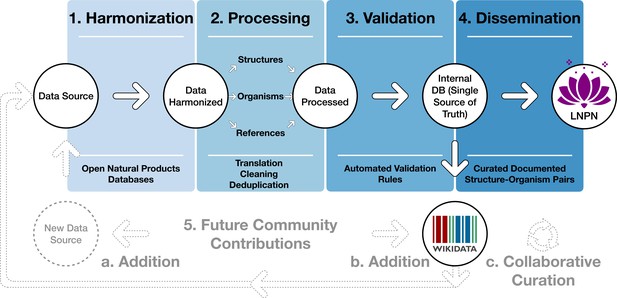
Blueprint of the LOTUS initiative.
Data undergo a four-stage process: (1) Harmonization, (2) Processing, (3) Validation, and (4) Dissemination. The process was designed to incorporate future contributions (5), either by the addition of new data from within Wikidata (a) or new sources (b) or via curation of existing data (c). The figure is available under the CC0 license at https://commons.wikimedia.org/wiki/File:Lotus_initiative_1_blueprint.svg.
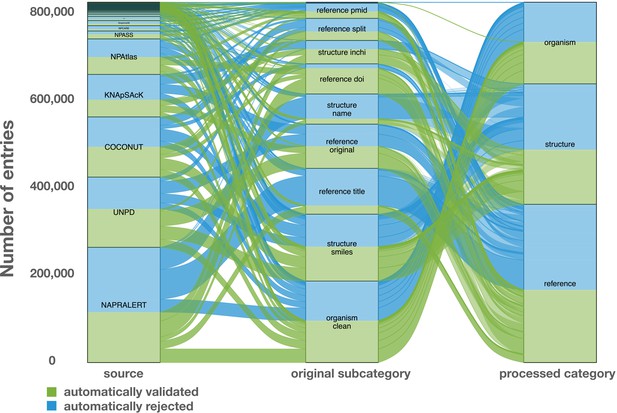
Alluvial plot of the data transformation flow within LOTUS during the automated curation and validation processes.
The figure also reflects the relative proportions of the data stream in terms of the contributions from the various sources (‘source’ block, left), the composition of the harmonized subcategories (‘original subcategory’ block, middle) and the validated data after curation (‘processed category’ block, right). Automatically validated entries are represented in green, rejected entries in blue. The figure is available under the CC0 license at https://commons.wikimedia.org/wiki/File:Lotus_initiative_1_alluvial_plot.svg.

Illustration of the ‘found in taxon’ statement section on the Wikidata page of erysodine Q27265641 showing a selection of erysodine-containing taxa and the references documenting these occurrences.
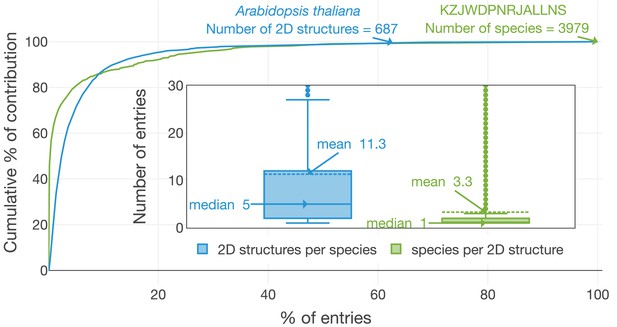
Distribution of ‘structures per organism’ and ‘organisms per structure’.
The number of organisms linked to the planar structure of β-sitosterol (KZJWDPNRJALLNS) and the number of chemical structures in Arabidopsis thaliana are two exemplary highlights. A. thaliana contains 687 different short InChIKeys (i.e. 2D structures) and KZJWDPNRJALLNS is reported in 3979 distinct organisms. Less than 10% of the species contain more than 80% of the structural diversity present within LOTUS. In parallel, 80% of the species present in LOTUS are linked to less than 10% of the structures. The figure is available under the CC0 license at https://commons.wikimedia.org/wiki/File:Lotus_initiative_1_structure_organism_distribution.svg.
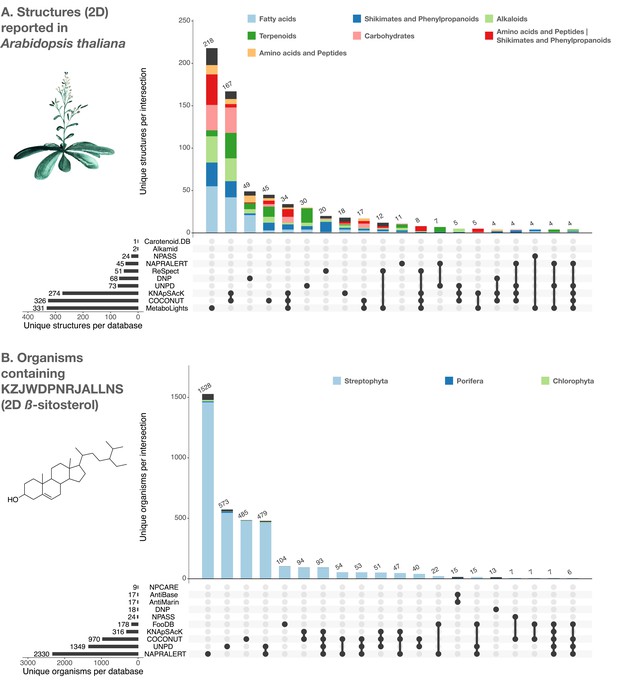
UpSet plots of the individual contribution of electronic NP resources to the planar structures found in Arabidopsis thaliana (A) and to organisms reported to contain the planar structure of β-sitosterol (KZJWDPNRJALLNS) (B).
UpSet plots are evolved Venn diagrams, allowing to represent intersections between multiple sets. The horizontal bars on the lower left represent the number of corresponding entries per electronic NP resource. The dots and their connecting line represent the intersection between source and consolidate sets. The vertical bars indicate the number of entries at the intersection. For example, 479 organisms containing the planar structure of β-sitosterol are present in both UNPD and NAPRALERT, whereas each of them respectively reports 1349 and 2330 organisms containing the planar structure of β-sitosterol. The figure is available under the CC0 license at https://commons.wikimedia.org/wiki/File:Lotus_initiative_1_upset_plot.svg.
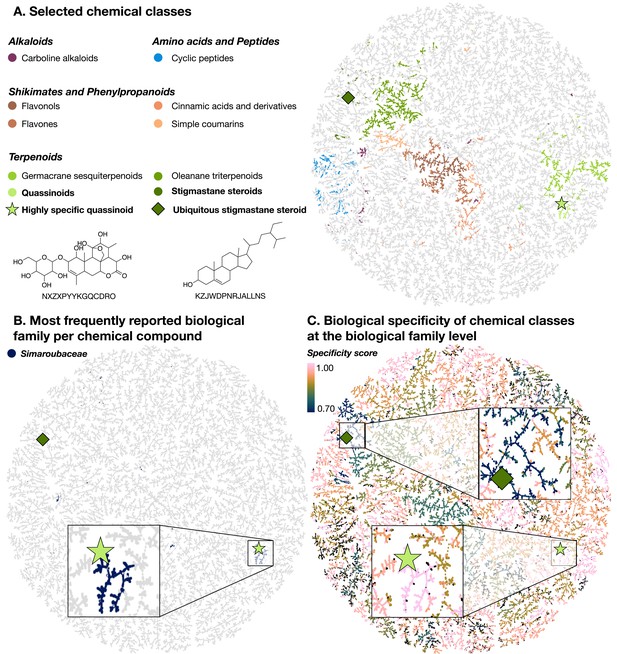
TMAP visualizations of the chemical diversity present in LOTUS.
Each dot corresponds to a chemical structure. A highly specific quassinoid (NXZXPYYKGQCDRO) (light green star) and an ubiquitous stigmastane steroid (KZJWDPNRJALLNS) (dark green diamond) are mapped as examples in all visualizations. In panel A., compounds (dots) are colored according to the NPClassifier chemical class they belong to. In panel B., compounds that are mostly reported in the Simaroubaceae family are highlighted in blue. Finally, in panel C., the compounds are colored according to the specificity score of chemical classes found in biological organisms. This biological specificity score at a given taxonomic level for a given chemical class is calculated as a Jensen-Shannon divergence. A score of 1 suggests that compounds are highly specific, 0 that they are ubiquitous. Zooms on a group of compounds of high biological specificity score (in pink) and on compounds of low specificity (blue) are depicted. An interactive HTML visualization of the LOTUS TMAP is available at https://lotus.nprod.net/post/lotus-tmap/ and archived at https://doi.org/10.5281/zenodo.5801807 (Rutz and Gaudry, 2021). The figure is available under the CC0 license at https://commons.wikimedia.org/wiki/File:Lotus_initiative_1_biologically_interpreted_chemical_tree.svg.
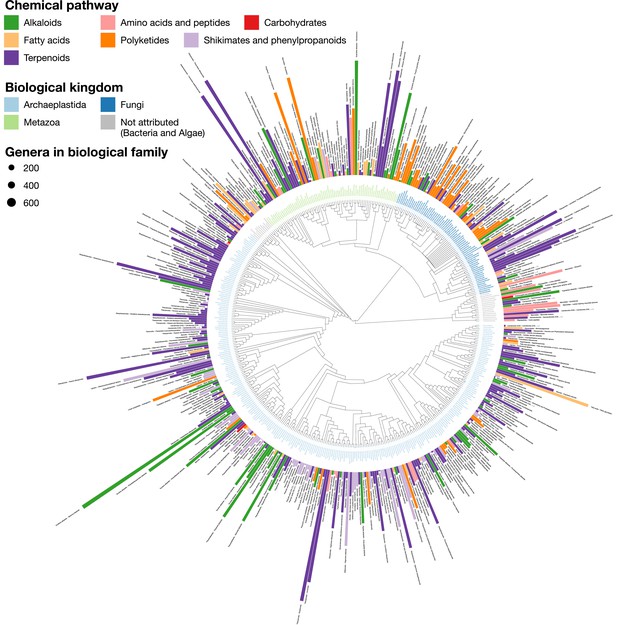
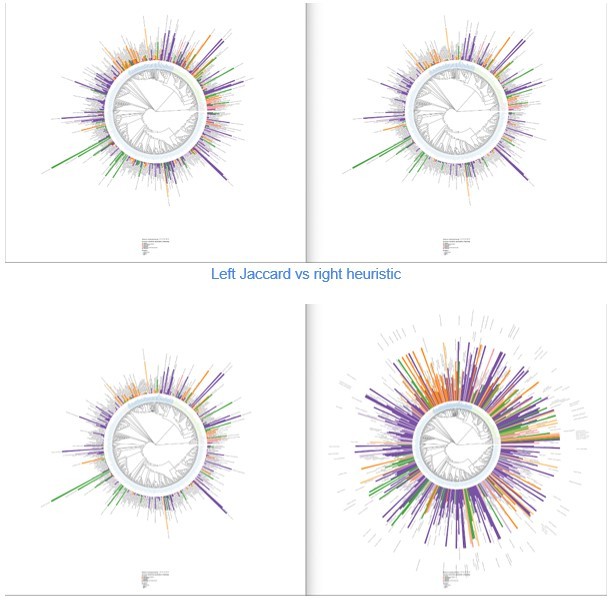
LOTUS provides new means of exploring and representing chemical and biological diversity.
The tree generated from current LOTUS data builds on biological taxonomy and employs the kingdom as tips label color (only families containing 50+ chemical structures were considered). The outer bars correspond to the most specific chemical class found in the biological family. The height of the bar is proportional to a specificity score corresponding to a Jaccard index between the chemical class and the biological family. The bar color corresponds to the chemical pathway of the most specific chemical class in the NPClassifier classification system. The size of the leaf nodes corresponds to the number of genera reported in the family. The figure is vectorized and zoomable for detailed inspection and is available under the CC0 license at https://commons.wikimedia.org/wiki/File:Lotus_initiative_1_chemically_interpreted_biological_tree.svg.
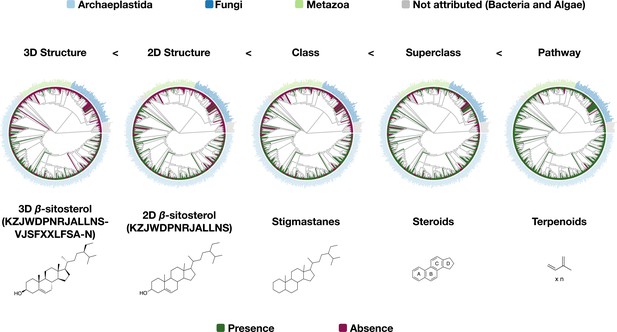
Distribution of β-sitosterol and related chemical parents among families with at least 50 reported compounds present in LOTUS.
Script used for the generation of each tree in the figure is the same (src/4_visualizing/plot_magicTree.R) as for Figure 7 as both figures are related. The figure is available under CC0 license at https://commons.wikimedia.org/wiki/File:Lotus_initiative_1_chemically_interpreted_biological_tree_supplement.svg.
Tables
Example of a referenced structure-organism pair before and after curation.
| Structure | Organism | Reference | |
|---|---|---|---|
| Before curation | Cyathocaline | Stem bark of Cyathocalyx zeylanica CHAMP. ex HOOK. f. & THOMS. (Annonaceae) | Wijeratne E. M. K., de Silva L. B., Kikuchi T., Tezuka Y., Gunatilaka A. A. L., Kingston D. G. I., J. Nat. Prod., 58, 459–462 (1995). |
| After curation | VFIIVOHWCNHINZ-UHFFFAOYSA-N | Cyathocalyx zeylanicus | 10.1021 /NP50117A020 |
Potential questions about structure-organism relationships and corresponding Wikidata queries.
| Question | Wikidata SPARQL query |
|---|---|
| What are the compounds present in Mouse-ear cress (Arabidopsis thaliana) or its child taxa? | https://w.wiki/4Vcv |
| Which organisms are known to contain β-sitosterol? | https://w.wiki/4VFn |
| Which organisms are known to contain stereoisomers of β-sitosterol? | https://w.wiki/4VFq |
| Which pigments are found in which taxa, according to which reference? | https://w.wiki/4VFx |
| What are examples of organisms where compounds were found in an organism sharing the same parent taxon, but not in the organism itself? | https://w.wiki/4Wt3 |
| Which Zephyranthes species lack compounds known from at least two species in the genus? | https://w.wiki/4VG3 |
| How many compounds are structurally similar to compounds labeled as antibiotics? (grouped by the parent taxon of the containing organism) | https://w.wiki/4VG4 |
| Which organisms contain indolic scaffolds? Count occurrences, group and order the results by the parent taxon. | https://w.wiki/4VG9 |
| Which compounds with known bioactivities were isolated from Actinobacteria, between 2014 and 2019, with related organisms and references? | https://w.wiki/4VGC |
| Which compounds labeled as terpenoids were found in Aspergillus species, between 2010 and 2020, with related references? | https://w.wiki/4VGD |
| Which are the available referenced structure-organism pairs on Wikidata? (example limited to 1000 results) | https://w.wiki/4VFh |
Distribution and specificity of chemical structures across four important NP reservoirs: plants, fungi, animals, and bacteria.
When the chemical structure/class appeared only in one group and not the three others, they were counted as ‘specific’. Chemical classes were attributed with NPClassifier.
| Group | Organisms | 2D Structure-Organism pairs | 2D chemical structures | Specific 2D chemical structures | Chemical classes | Specific chemical classes |
|---|---|---|---|---|---|---|
| Plantae | 28,439 | 342,891 | 95,191 | 90,672 (95%) | 545 | 59 (11%) |
| Fungi | 4,003 | 36,950 | 22,594 | 20,194 (89%) | 417 | 19 (5%) |
| Animalia | 2,716 | 24,114 | 15,242 | 11,822 (78%) | 455 | 14 (3%) |
| Bacteria | 1,555 | 23,198 | 15,895 | 14,130 (89%) | 385 | 43 (11%) |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Software, algorithm | Lotus-processor code | This work (https://github.com/lotusnprod/lotus-processor, Rutz, 2022a) | Archived at https://doi.org/10.5281/zenodo.5802107 | |
| Software, algorithm | Lotus-web code | This work (https://github.com/lotusnprod/lotus-web, Rutz, 2022b) | Archived at https://doi.org/10.5281/zenodo.5802119 | |
| Software, algorithm | Lotus-wikidata-interact code | This work (https://github.com/lotusnprod/lotus-wikidata-interact, Rutz, 2022c) | Archived at https://doi.org/10.5281/zenodo.5802113 | |
| Software, algorithm | Global Names Architeture | https://globalnames.org | QID:Q65691453 | See Additional executable files |
| Software, algorithm | Java | https://www.java.com | QID:Q251 | |
| Software, algorithm | Kotlin | https://kotlinlang.org | QID:Q3816639 | See Kotlin packages |
| Software, algorithm | Manubot | https://manubot.org | QID:Q96473455 RRID:SCR_018553 | Repository available at https://github.com/lotusnprod/lotus-manuscript |
| Software, algorithm | NPClassifier | https://npclassifier.ucsd.edu | See https://doi.org/10.1021/acs.jnatprod.1c00399 | |
| Software, algorithm | OPSIN | https://github.com/dan2097/opsin | QID:Q26481302 | See Additional executable files |
| Software, algorithm | Python Programming Language | https://www.python.org | QID:Q28865 RRID:SCR_008394 | See Python packages |
| Software, algorithm | R Project for Statistical Computing | https://www.r-project.org | QID:Q206904 RRID:SCR_001905 | See R packages |
| Software, algorithm | Molconvert | https://docs.chemaxon.com/display/docs/molconvert.md | QID:Q55377678 | See Chemical structures |
| Software, algorithm | Wikidata | https://www.wikidata.org | QID:Q2013 RRID:SCR_018492 | Project page https://www.wikidata.org/wiki/Wikidata:WikiProject_Chemistry/Natural_products |
| Other | Lotus custom dictionaries | This work | Archived at https://doi.org/10.5281/zenodo.5801798 | |
| Other | Chemical identifier resolver | https://cactus.nci.nih.gov/chemical/structure | See Chemical structures | |
| Other | CrossRef | https://www.crossref.org | QID:Q5188229 RRID:SCR_003217 | See References |
| Other | PubChem | https://pubchem.ncbi.nlm.nih.gov | QID:Q278487 RRID:SCR_004284 | LOTUS data https://pubchem.ncbi.nlm.nih.gov/source/25132 |
| Other | PubMed | https://pubmed.ncbi.nlm.nih.gov | QID:Q180686 RRID:SCR_004846 | See References |
| Other | Taxonomic data sources | https://resolver.globalnames.org/data_sources | See Translation | |
| Other | Natural Products data sources | See Appendix 1 |
Data sources list.
| Database | Type | Initial retrieved unique entries | Cleaned referenced structure-organism pairs | Pairs validated for Wikidata export | Actual validated pairs on Wikidata | Website | Article | Retrieval | License status | Contact | Dump | Status |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| afrotryp | open | 313 | 93 | 55 | 54 | - | article (Ibezim et al., 2017) | download | license_copyright | Fidele Ntie-Kang or Ngozi Justina Nwodo | YES | unmaintained |
| alkamid | open | 4,416 | 2,639 | 2,309 | 2,160 | website | article (Boonen et al., 2012) | script | license_copyright | Bart De Spiegeleer | NO | maintained |
| antibase | commercial | 46,956 | 45,221 | - | - | - | - | - | - | - | NO | unmaintained |
| antimarin | commercial | 73,017 | 67,559 | - | - | - | - | - | - | - | NO | unmaintained |
| biofacquim | open | 531 | 519 | 519 | 511 | website (old version) | article_old; article_new (Pilón-Jiménez et al., 2019) | download | license_CCBY_4.0 | José Medina-Franco | YES | maintained |
| biophytmol | open | 543 | 558 | 322 | 308 | website | article (Sharma et al., 2014) | script | license_CCBY | Anshu Bhardwaj | NO | unmaintained |
| carotenoiddb | open | 2,922 | 639 | 530 | 485 | website | article (Yabuzaki, 2017) | script | license_copyright | yzjunko@gmail.com | NO | maintained |
| coconut | open | 5,757,872 | 5,723,691 | 153,981 | 140,877 | website | article (Sorokina and Steinbeck, 2020b) | download | license_CCBY_4.0 | Maria Sorokina | YES | maintained |
| cyanometdb | open | 1,905 | 1,631 | 1,621 | 1,605 | - | article (Jones et al., 2021) | download | license_CCBY_4.0 | elisabeth.janssen@eawag.ch | YES | maintained |
| datawarrior | open | 589 | 541 | 71 | 60 | website | article (Sander et al., 2015) | download | no_license | thomas.sander@idorsia.com | YES | retired |
| dianatdb | open | 290 | 323 | 115 | 111 | website | article (Madariaga-Mazón et al., 2021) | download | license_CCBY_NC | amadariaga@iquimica.unam.mx or kmtzm@unam.mx | YES | maintained |
| dnp | commercial | 205,072 | 254,573 | - | - | website | - | script | - | support@taylorfrancis.com | NO | maintained |
| drduke | open | 90,675 | 9,660 | 6,184 | 5,222 | website | - | download | license_CC0 | agref@usda.gov | YES | maintained |
| foodb | restricted | 81,941 | 39,662 | - | - | website | - | download | license_CCBY_NC | jreid3@ualberta.ca (Jennifer) | YES | unmaintained |
| inflamnat | open | 665 | 632 | 306 | 268 | - | article (Zhang et al., 2019) | download | license_copyright | xiaoweilie@ynu.edu.cn | YES | unmaintained |
| knapsack | open | 132,127 | 139,336 | 59,945 | 55,186 | website | article (Shinbo et al., 2006) | script | license_copyright | skanaya@gtc.naist.jp | NO | maintained |
| metabolights | open | 38,208 | 37,704 | 6,241 | 5,687 | website | article (Haug et al., 2020) | download | license_copyright | - | YES | maintained |
| mibig | open | 1,310 | 1,139 | 638 | 535 | website | article (Kautsar et al., 2020) | download | license_CCBY_4.0 | Tilmann Weber orMarnix Medema | YES | unmaintained |
| mitishamba | open | 1,071 | 534 | 294 | 291 | website | article (Derese et al., 2019) | script | license_copyright | - | NO | defunct |
| nanpdb | open | 5,752 | 6,383 | 5,937 | 5,283 | website | article (Ntie-Kang et al., 2017) | script | license_copyright | ntiekfidele@gmail.com stefan.guenther@pharmazie.uni-freiburg.de | NO | maintained |
| napralert | commercial | 681,401 | 392,498 | 294,818 | 270,743 | website | article (Graham and Farnsworth, 2010) | - | license_copyright | napralert@uic.edu | NO | defunct |
| npass | open | 290,535 | 30,185 | 25,429 | 23,612 | website | article (Zeng et al., 2018) | download | license_CCBY_NC | phacyz@nus.edu.sg jiangyy@sz.tsinghua.edu.cn iaochen@163.com | YES | unmaintained |
| npatlas | open | 32,539 | 34,726 | 34,548 | 33,087 | website | article (van Santen et al., 2019; ) | download | license_CCBY_4.0 | rliningt@sfu.ca | YES | maintained |
| npcare | open | 7,763 | 5,878 | 3,790 | 3,538 | website | article (Choi et al., 2017) | download | license_CCBY_4.0 | choihwanho@gmail.com | YES | unmaintained |
| npedia | open | 82 | 99 | 28 | 28 | website | article (Tomiki et al., 2006) | script | no_license | hisyo@riken.jp npd@riken.jp | NO | defunct |
| nubbe | open | 2,189 | 2,340 | 2,340 | 2,119 | website | article (Pilon et al., 2017) | - | license_copyright | Vanderlan S. Bolzani | NO | maintained |
| pamdb | open | 3,046 | 2,820 | 24 | 24 | website | article (Huang et al., 2018) | download | license_CCBY_NC | awilks@rx.umaryland.edu aoglesby@rx.umaryland.edu mkane@rx.umaryland.edu | YES | unmaintained |
| phenolexplorer | open | 8,077 | 8,700 | 7,123 | 5,721 | website | article (Rothwell et al., 2013) | download | license_copyright | scalberta@iarc.fr | YES | unmaintained |
| phytohub | open | 2,349 | 1,145 | 132 | 94 | website | article (Giacomoni et al., 2017) | script | no_license | claudine.manach@inra.fr | YES | unmaintained |
| procardb | open | 6,556 | 6,278 | 60 | 55 | website | article (Nupur et al., 2016) | script | license_CCBY_4.0 | Anil Kumar PinnakaAshwani Kumar | NO | unmaintained |
| respect | open | 2,759 | 1,064 | 634 | 547 | website | article (Sawada et al., 2012) | download | license_CCBY_NC_2.1_Japan | ksaito@psc.riken.jp | YES | unmaintained |
| sancdb | open | 860 | 925 | 747 | 732 | website | article (Hatherley et al., 2015) | script | license_CCBY_4.0 | Özlem Tastan Bishop | NO | unmaintained |
| streptomedb | open | 71,638 | 33,217 | 20,715 | 18,395 | website | article (Klementz et al., 2016) | download | license_copyright | stefan.guenther@pharmazie.uni-freiburg.de | YES | maintained |
| swmd | open | 1,075 | 1,751 | 1,597 | 1,479 | website | article (Davis and Vasanthi, 2011) | script | license_CCBY_4.0 | Dicky.John@gmail.com | NO | unmaintained |
| tmdb | open | 2,116 | 533 | 26 | 24 | website | article (Yue et al., 2014) | script | license_copyright | Xiao-Chun WanGuan-Hu Bao | NO | unmaintained |
| tmmc | open | 15,033 | 7,833 | 5,826 | 4,015 | website | article (Kim et al., 2015a) | download | license_copyright | Jeong-Ju Lee | YES | unmaintained |
| tppt | open | 27,182 | 23,872 | 684 | 641 | website | article (Günthardt et al., 2018) | download | license_copyright | thomas.bucheli@agroscope.admin.ch | YES | unmaintained |
| unpd | open | 331,242 | 304,683 | 211,158 | 197,710 | website | article (Gu et al., 2013) | - | license_CCBY_4.0 | lirongc@pku.edu.cn xiaojxu@pku.edu.cn | NO | defunct |
| wakankensaku | open | 367 | 224 | 208 | 202 | website | - | script | - | - | NO | defunct |
| Wikidata | open | 951,268 | 960,611 | 959,747 | 919,752 | website | - | download | license_CC0 | - | YES | maintained |
Summary of the Validation Statistics.
| First validation dataset (n = 420) | Second validation dataset (n = 100) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Reference Type | True positive | False positive | False negative | True negative | Ratio | Precision | Recall | F0.5 score | True positive | False negative |
| Original | 80 | 6 | 7 | 11 | 0.31 | 0.93 | 0.92 | 0.92 | 38 | 1 |
| Pubmed | 37 | 1 | 5 | 6 | 0.30 | 0.97 | 0.88 | 0.92 | 5 | 1 |
| DOI | 115 | 6 | 0 | 6 | 0.19 | 0.95 | 1.00 | 0.97 | 43 | 1 |
| Title | 38 | 2 | 0 | 16 | 0.12 | 0.95 | 1.00 | 0.97 | 7 | 0 |
| Split | 8 | 0 | 15 | 27 | 0.08 | 1.00 | 0.35 | 0.52 | 4 | 0 |
| Publishing details | 1 | 0 | 1 | 32 | 0.01 | 1.00 | 0.50 | 0.67 | 0 | 0 |
| Total | 279 | 15 | 28 | 98 | 1.00 | - | - | - | 97 | 3 |
| Corrected total | - | - | - | - | - | 0.96 | 0.89 | 0.91 | - | - |
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/70780/elife-70780-transrepform1-v1.pdf
-
Supplementary file 1
Wikidata Queries.
- https://cdn.elifesciences.org/articles/70780/elife-70780-supp1-v1.docx
-
Supplementary file 2
Wikidata Entry Creation Tutorial.
- https://cdn.elifesciences.org/articles/70780/elife-70780-supp2-v1.docx