Morphometric analysis of lungfish endocasts elucidates early dipnoan palaeoneurological evolution
Figures
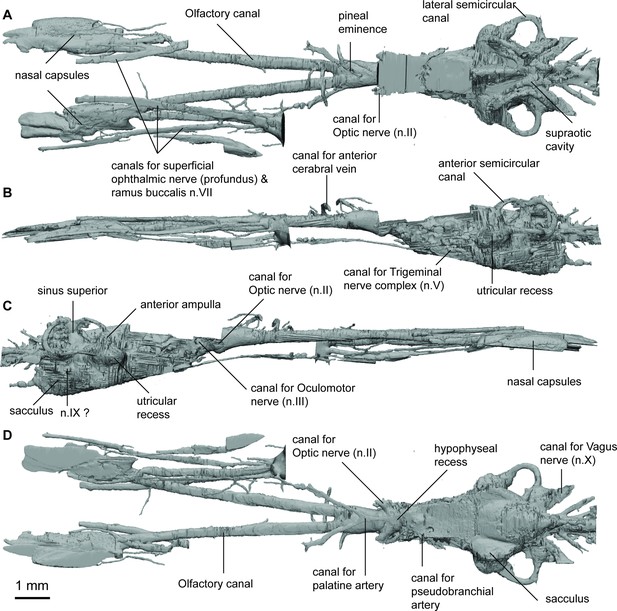
Endocast of Griphognathus whitei (NHMUK PV P56054) in (a), dorsal; (b, c), lateral; (d), ventral views.
Endocast of Iowadipterus halli (FMNH PF 12323) in (a), dorsal; (b), lateral left; (c), lateral right; and (d), ventral views.
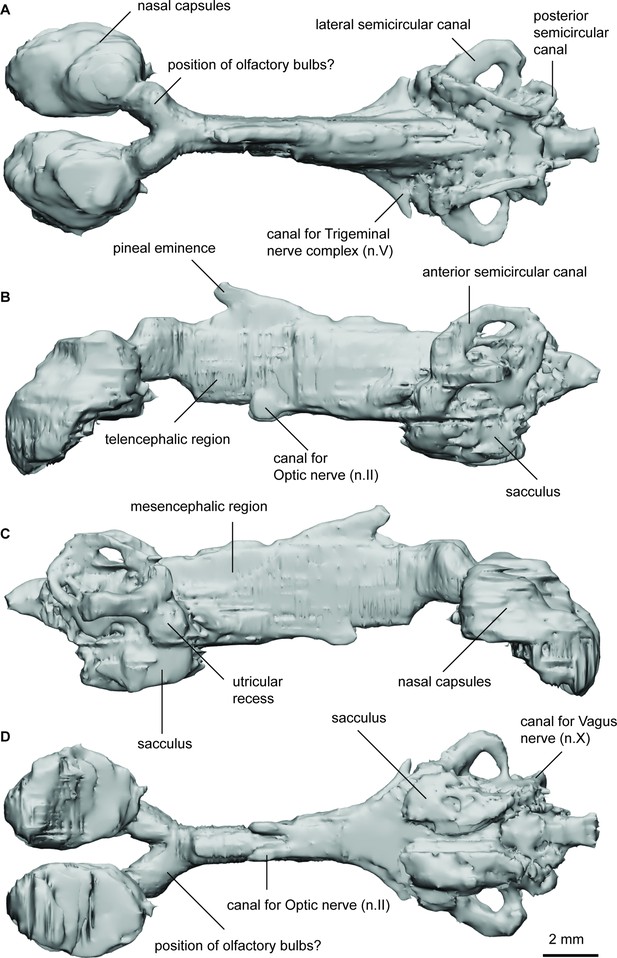
Endocast of Orlovichthys limnatis (PIN 3725/110) in (a), dorsal; (b), lateral left; (c), lateral right; and (d), ventral views.
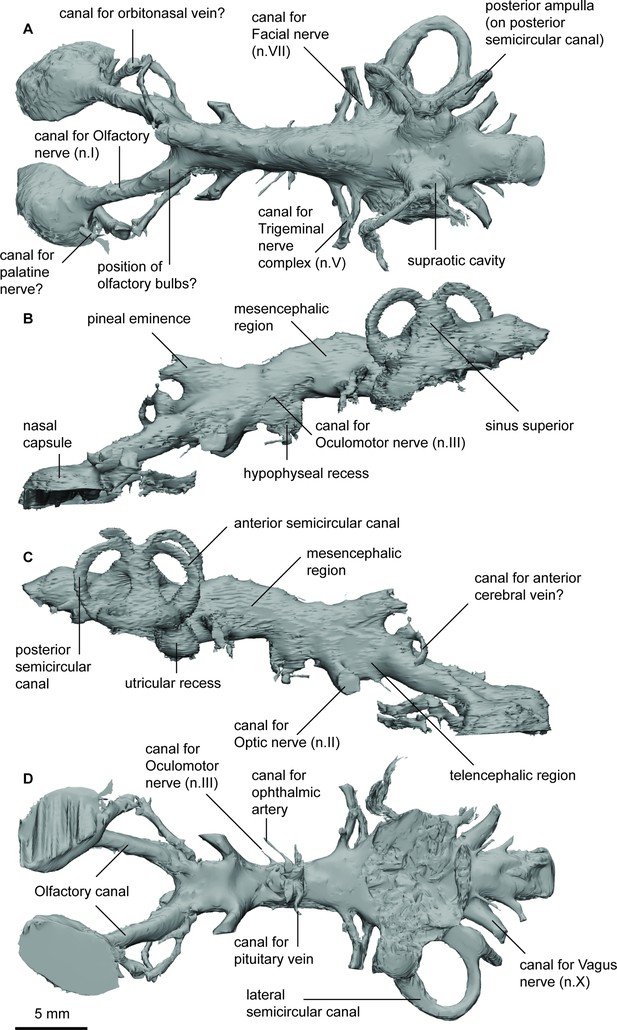
Endocast of Pillararhynchus longi (ANU 49196) in (a), dorsal; (b), lateral left; (c), lateral right; and (d), ventral views.
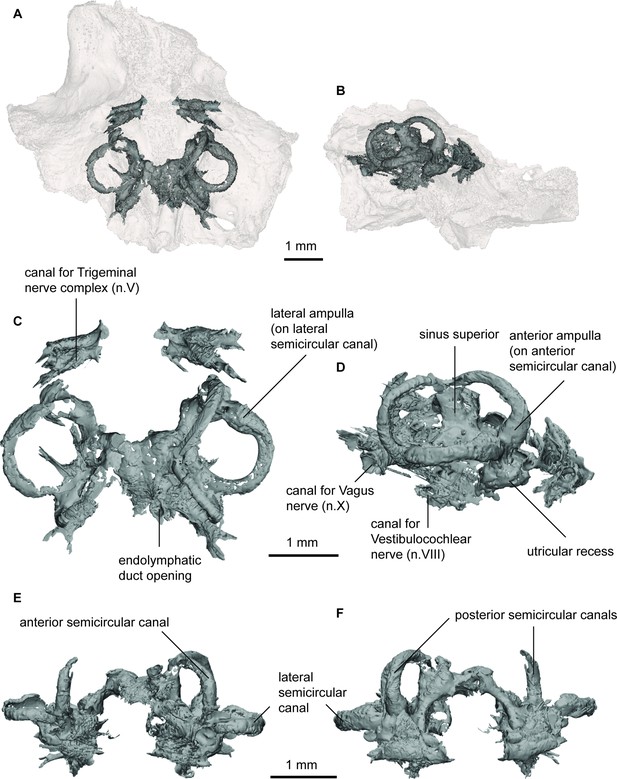
Endocast of Gogodipterus paddyensis (WAM 70.4.250) labyrinth region in (a), dorsal; and (b), right lateral views with transparent skull overlay; (c), dorsal; (d), right lateral; (e), anterior; (f), posterior views of labyrinth endocast only.
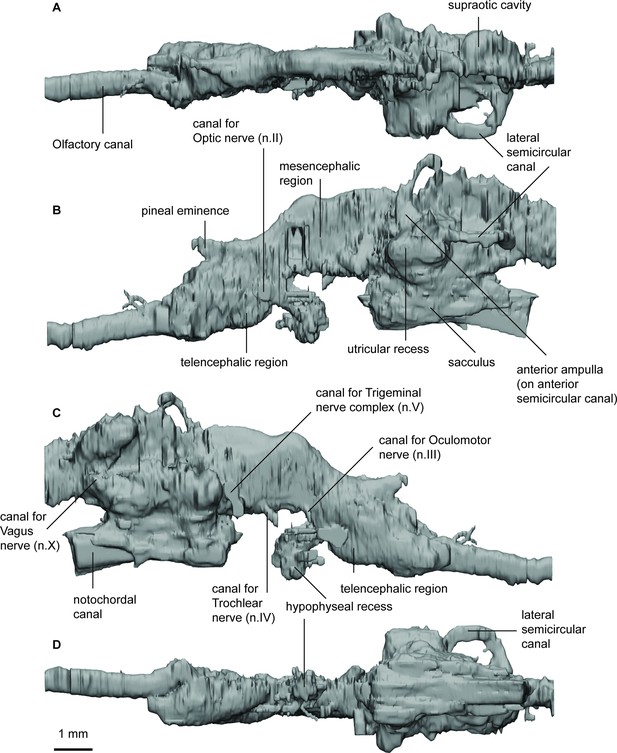
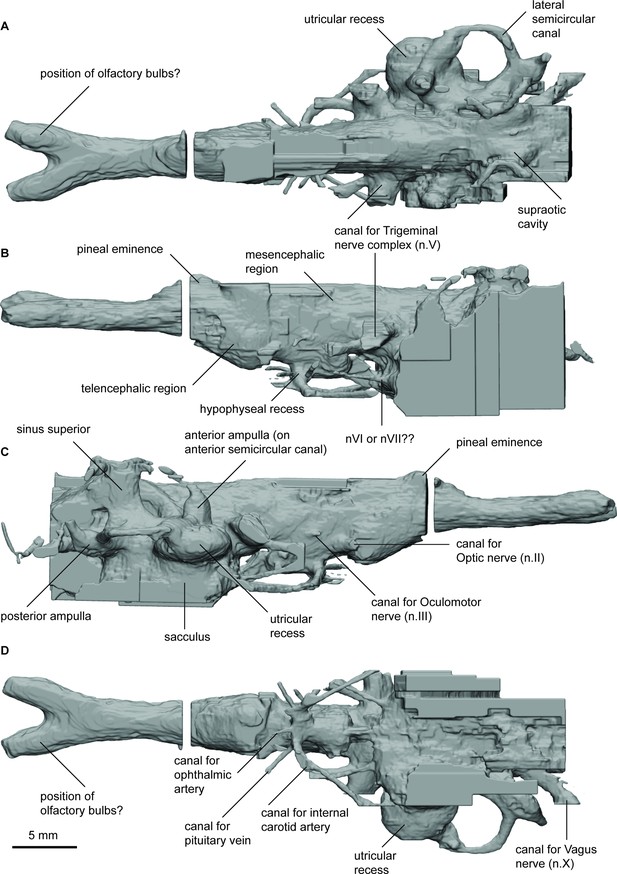
Endocast of Rhinodipterus ulrichi (NRM P6609a) in (a), dorsal; (b), lateral left; (c), lateral right; and (d), ventral views.
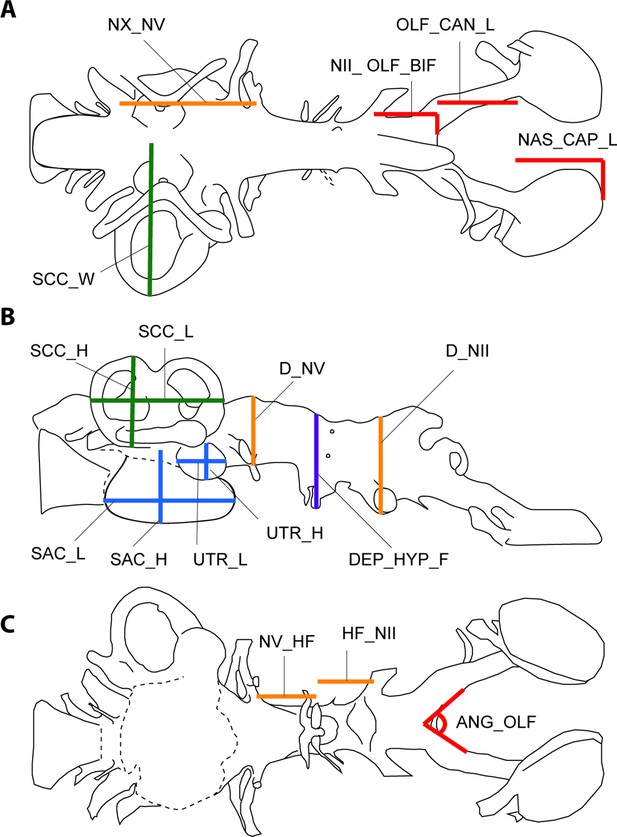
Illustration of 17 endocast variables measured on endocast in (a), dorsal; (b), left lateral; and (c), ventral view.
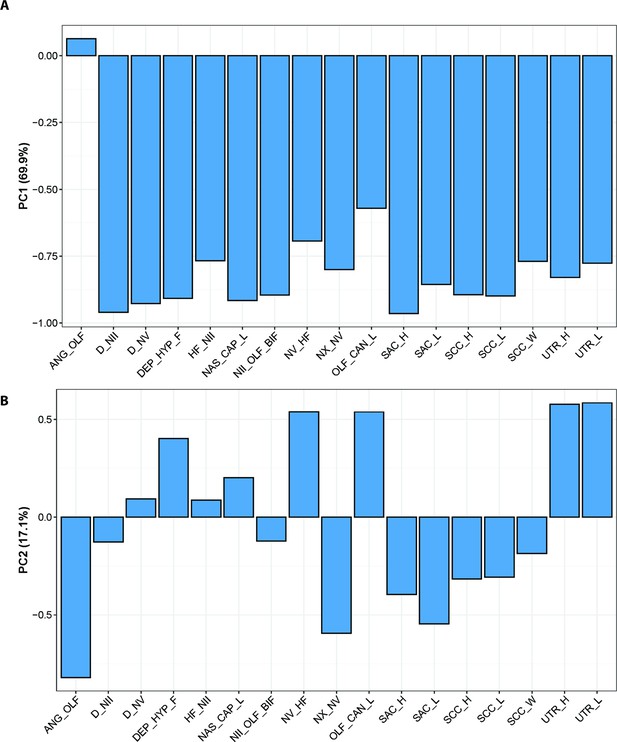
ANG_OLF, angle of bifurcation between olfactory canals; D_NII, depth of cranial endocast at level of n.II; D_NV, depth of cranial endocast at level of n.V; DEP_HYP_F, depth of cranial endocast at level of hypophyseal fossa; HF_NII, distance from hypophyseal fossa to n.II; NAS_CAP_L, length of nasal capsules; NII_ OLF_BIF, distance from n.II to point of bifurcation between olfactory canals; NV_HF, distance from n.V to hypophyseal fossa; NX_NV, distance from n.V to n.X; OLF_CAN_L, length of olfactory canals from nasal capsules to point of bifurcation; SAC_H, height of sacculus; SAC_L, length of sacculus; SCC_H, height of semicircular canals; SCC_L, length of semicircular canals from anterior to posterior; SCC_W, width of semicircular canals from midline to lateral canal; UTR_H, height of utriculus; UTR_L, length of utriculus.
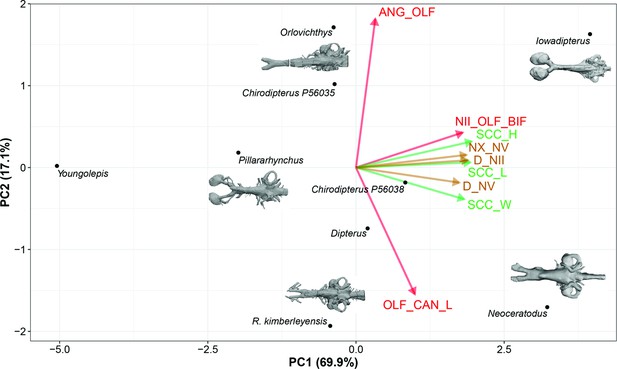
Standard Principal Component Analysis (PCA) biplot of lungfish endocast shape optimised for maximum number of included TAXA, PC1 vs PC2.
Loadings and eigenvalues shown in Figure 8—figure supplement 1.
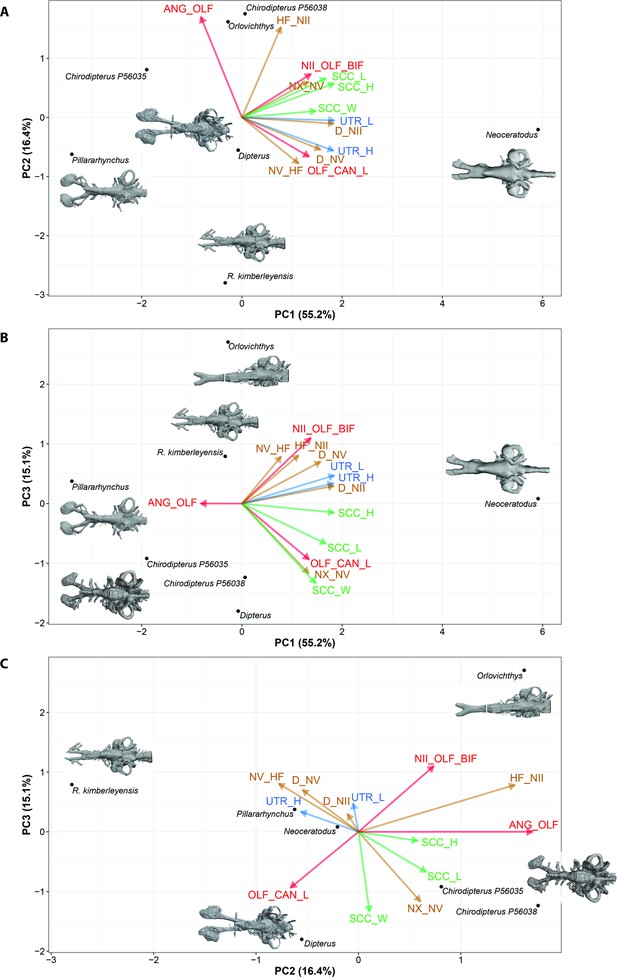
Standard Principal Component Analysis (PCA) biplot of lungfish endocast shape optimised for maximum number of included CHARACTERS.
(A), PC1 vs PC2; (B), PC1 vs PC3; (C), PC2 vs PC3. Loadings and eigenvalues shown in Figure 9—figure supplements 1 and 2.
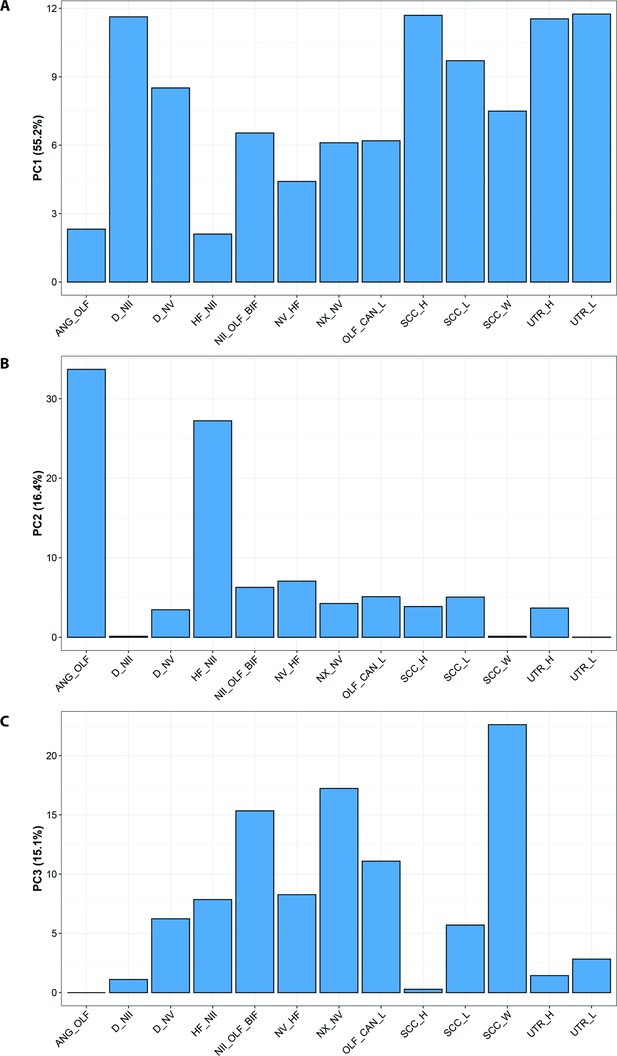
Standard PCA loadings optimised for CHARACTERS A, PC1; B, PC2; and C, PC3.
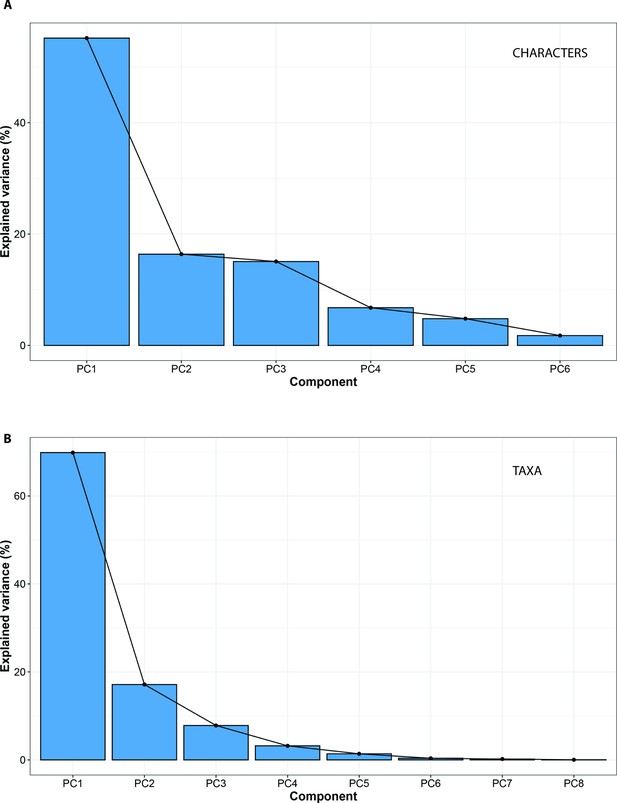
Standard PCA eigenvalues optimised for maximum A, CHARACTERS; and B, TAXA.
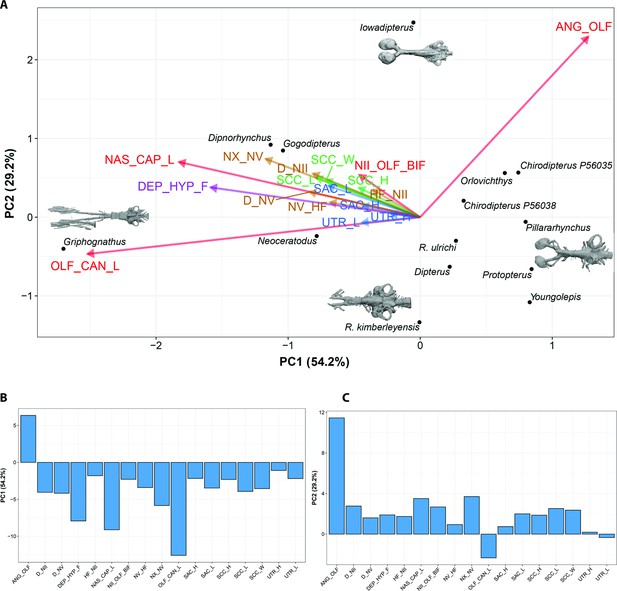
Bayesian Principle Component Analysis (BPCA) biplot of lungfish endocast shape (A), PC 1 vs PC 2 with endocasts of select taxa shown; and (B), (C) loadings for PCA 1–2.
Eigenvalues shown in Figure 10—figure supplement 1.
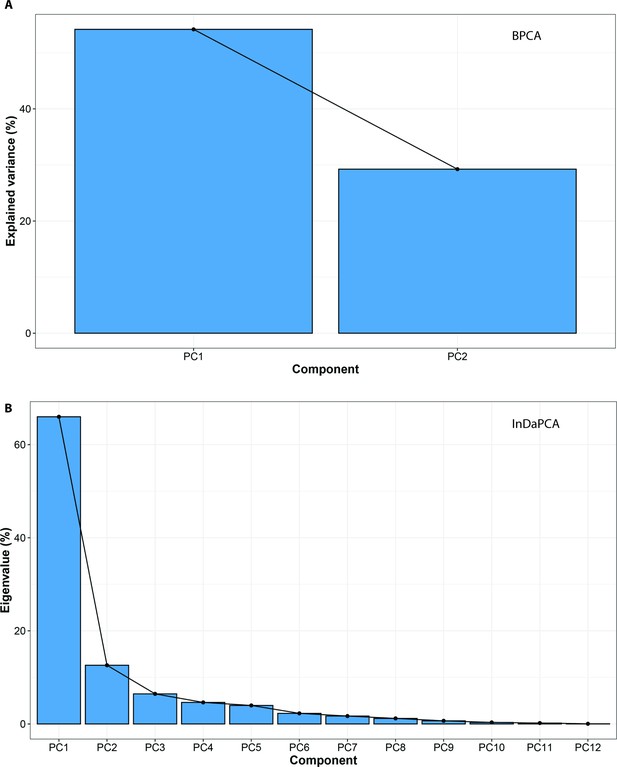
PCA eigenvalues for A, Bayesian Principle Component Analysis (BPCA); and B, Principle Component Analysis for incomplete data (InDaPCA).
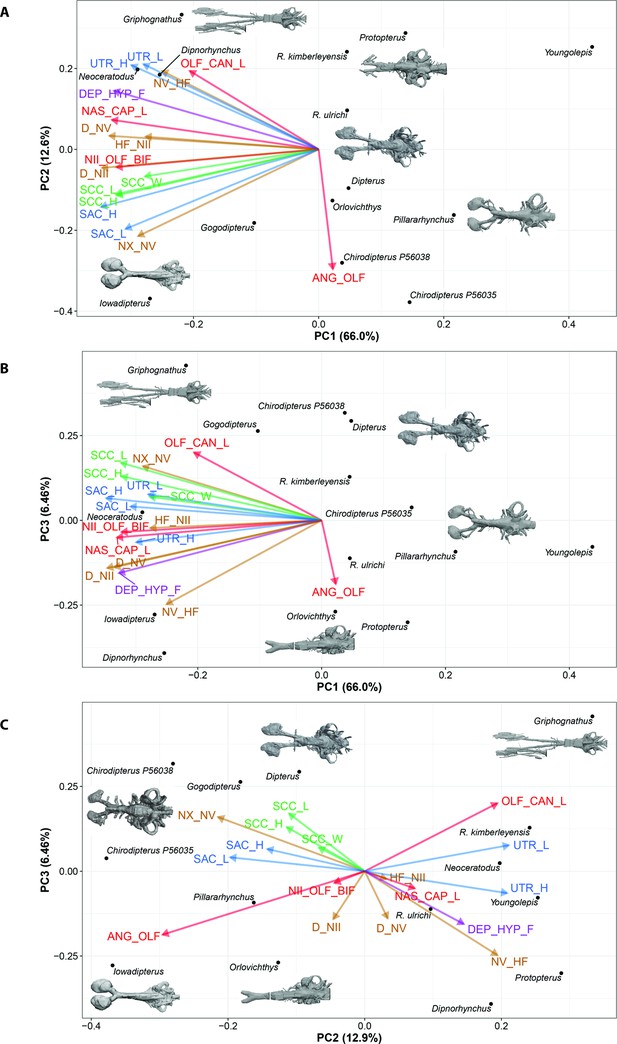
Principle Component Analysis for incomplete data (InDaPCA) biplot of lungfish endocast shape for (A), PC1 vs PC2; (B), PC1 vs PC3; (C), PC2 vs PC3, with endocasts of select taxa shown.
Explanation of variable abbreviations as per Figure 7.
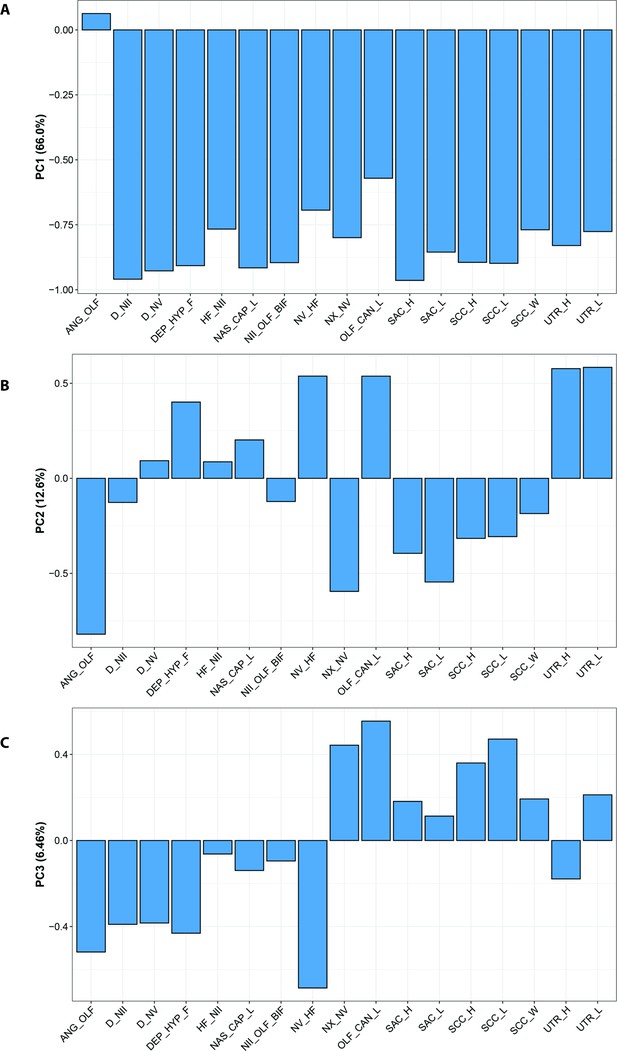
Principle Component Analysis for incomplete data (InDaPCA) loadings for A, PC1; B, PC2; and C, PC3.
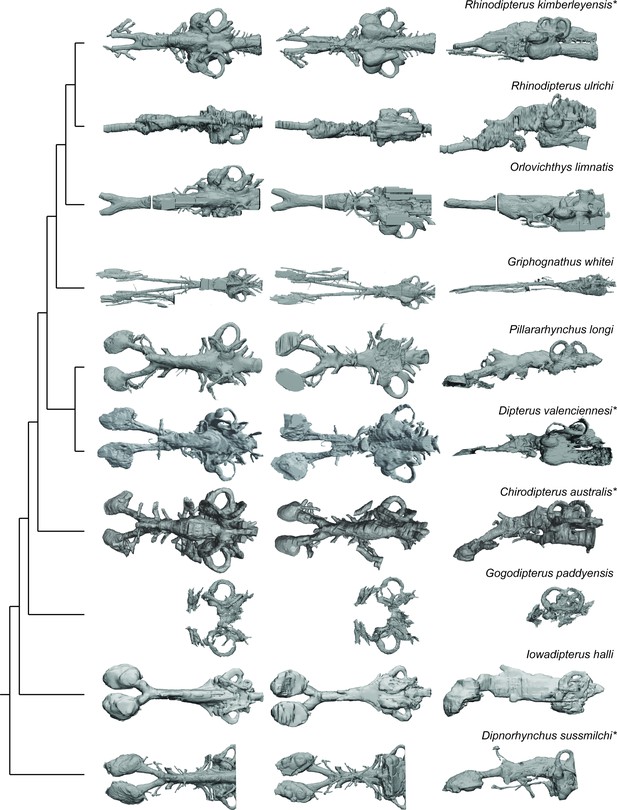
Palaeozoic lungfish endocasts in dorsal, ventral, and left lateral views, placed onto phylogeny adapted from Challands et al., 2019 .
*The following endocasts were first published elsewhere but new images were created for inclusion in this figure: Rhinodipterus kimberleyensis (Clement and Ahlberg, 2014) Figures 2 and 3; Dipterus valenciennesi (Challands, 2015) Figures 6 and 9; ‘Chirodipterus’ australis (Henderson and Challands, 2018) Figures 3–4; Dipnorhynchus sussmilchi (Clement et al., 2016a) Figures 2—4.
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/73461/elife-73461-transrepform1-v1.pdf
-
Source data 1
Table containing endocast measurements as a matrix, including additional comments and variables.
- https://cdn.elifesciences.org/articles/73461/elife-73461-data1-v1.xlsx
-
Source data 2
Table containing PCA loadings for the taxa-optimised analyses.
- https://cdn.elifesciences.org/articles/73461/elife-73461-data2-v1.xlsx
-
Source data 3
Table containing PCA loadings for the character-optimised analyses.
- https://cdn.elifesciences.org/articles/73461/elife-73461-data3-v1.xlsx
-
Source data 4
Table containing loadings for the BPCA.
- https://cdn.elifesciences.org/articles/73461/elife-73461-data4-v1.xlsx
-
Source data 5
Table containing loadings for the InDaPCA.
- https://cdn.elifesciences.org/articles/73461/elife-73461-data5-v1.xlsx