Common coupling map advances GPCR-G protein selectivity
Figures
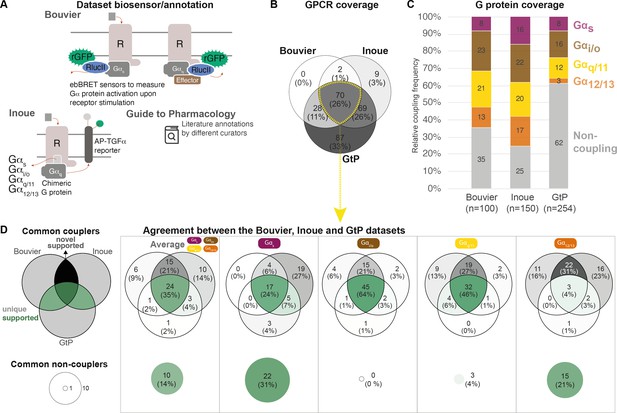
Coverage and agreement of the Bouvier lab, Inoue group, and Guide to Pharmacology (GtP) datasets.
(A) Biosensor principles used by the Bouvier and Inoue groups, and literature annotation stored in the GtP database. (B) Intersection of the G protein-coupled receptors (GPCRs) included in the Bouvier (n = 100), Inoue (n = 150), and GtP (n = 254) datasets. (C) Relative family distributions of G protein couplings across datasets. (D) Comparison of the G protein family coupling profiles of 70 GPCRs present in all of the Bouvier, Inoue, and GtP datasets. More detailed analysis of common G protein couplings for just the Bouvier and Inoue datasets is given in Figure 1—figure supplement 1. (A–D) Note: All analyses herein cover the 12 G proteins: Gs, Gi1, Gi2, GoA, GoB, Gz, Gq, G11, G14, G15, G12, and G13. Golf and Gi3 could not be analyzed, as they had not been tested by the Bouvier group. The Inoue data for the pairs Gi1-Gi2, GoA-GoB, and Gq-G11 were generated with identical chimera inserting the Gα C-terminal hexamer into a Gq backbone (Inoue et al., 2019) meaning that identical datapoints were used to assess their coverage and agreement with other datasets (Table 1).
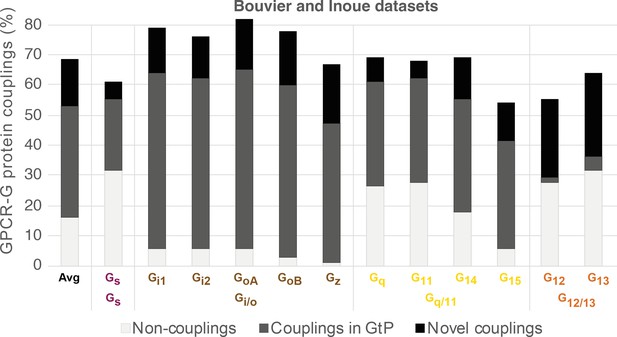
Common G protein-coupled receptor (GPCR-G) protein couplings in the Bouvier and Inoue datasets.
Black bars represent couplings not in Guide to Pharmacology (GtP) and therefore considered novel (novel couplings are listed in Figure 3). Frequencies are percent couplings among all receptors common to Bouvier and Inoue (the 100% includes noncoupling receptors). Gi3 and Golf (chimeras) are not included herein as they were only tested by the Inoue group. Inoue used the same chimeras to represent the pairs GoA-GoB, Gi1-Gi2, and Gq-G11 (Table 1).
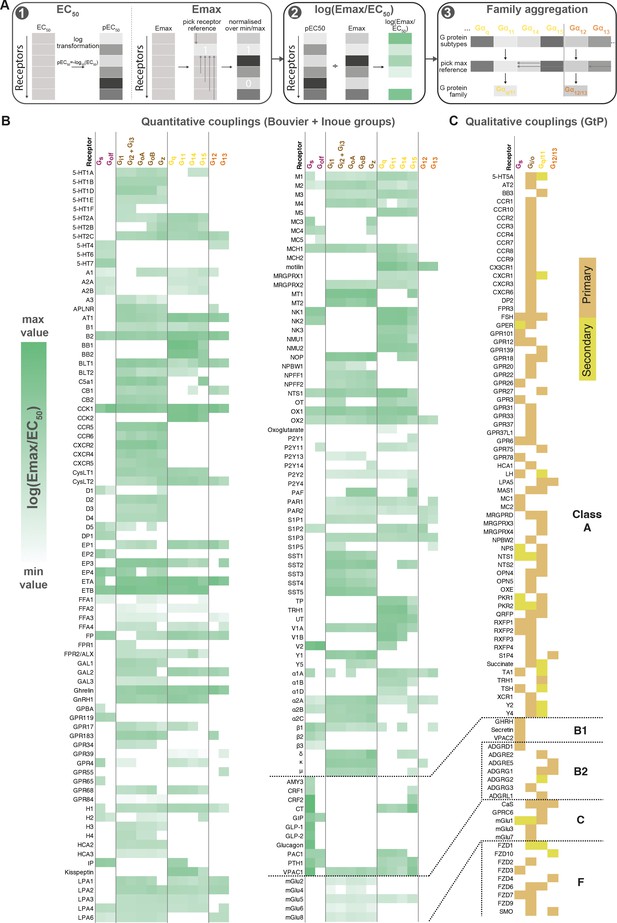
Map of G protein-coupled receptor (GPCR-G) protein couplings supported by at least two studies.
(A) Normalization approach. Emax minimum-maximum normalization, EC50 log transformation and use of log(Emax/EC50) as a combined measure with member-to-family aggregation by maximum G protein value. (B) Heatmap representation of log(Emax/EC50) values for 166 GPCRs tested by the Bouvier and/or Inoue labs (for couplings with dual data sources a mean is used). For Guide to Pharmacology (GtP), a G protein subtype is considered supported if the family has a known coupling. Gi2 and Gi3 are represented by the same/identical chimeric G protein in Inoue’s dataset (Table 1). (C) Heatmap representation of primary and secondary transducers for 90 GPCRs which couplings are only covered by the GtP database. (B–C) Empty cells (white) indicate no coupling. All source values are available in tab ‘Fig_4’ in Source data 5. Note: Researchers wishing to use this coupling map, optionally after applying own reliability criteria or cut-offs, can do so for any set of couplings in GproteinDb (Pándy-Szekeres et al., 2022).
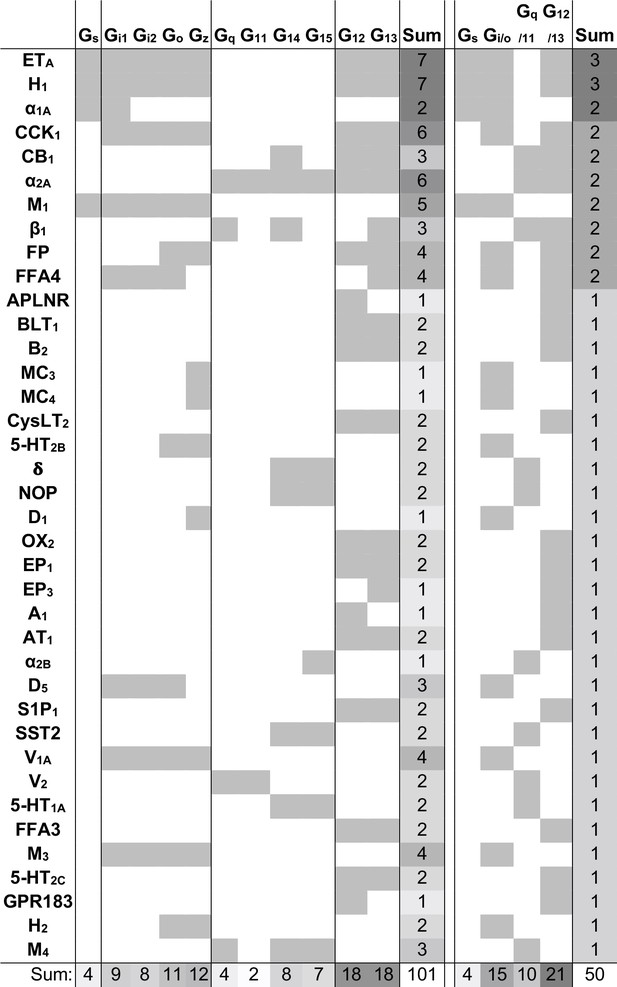
Novel G protein couplings identified by both the Bouvier and Inoue groups.
Novel couplings identified by the Bouvier and Inoue groups but not in Guide to Pharmacology. G protein families are here considered shared if at least one specific subtype is found to couple in both dataset, which is not the case for the bottommost four receptors.
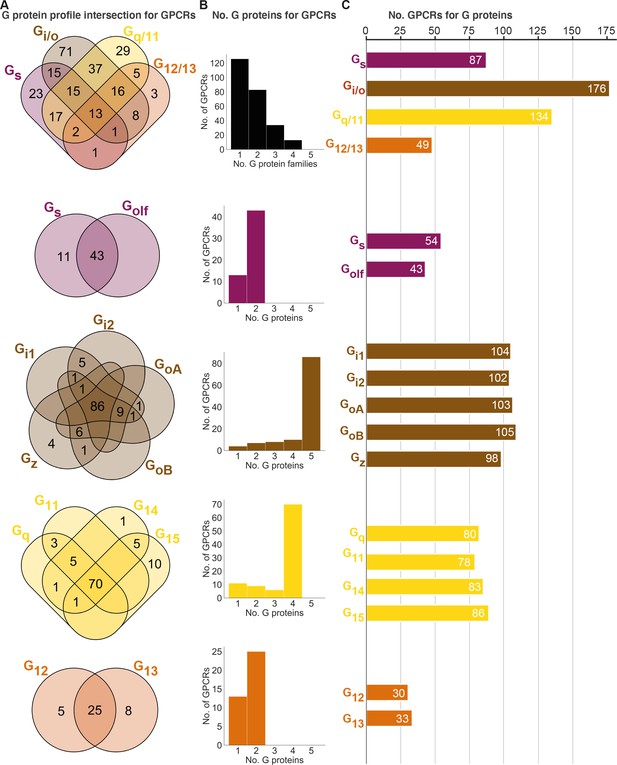
Unifying the three datasets reveals a large diversity in G protein-coupled receptor (GPCR) coupling selectivity.
(A) GPCR-G protein selectivity on the G protein family (top) and subtype levels: Venn diagrams showing the numbers of shared and unique receptors. The 0 values (no receptors) are omitted for clarity. (B) Receptor coupling promiscuity: number of receptors that couple to 1–4 G protein families (top) and 1–5 subtypes. (C) G protein coupling promiscuity: number of receptors that couple to each G protein family (top) or subtype. (A–C) Panels B–C are based on the couplings from the Bouvier and Inoue groups that are also supported by a second dataset and panel A additionally includes GtP couplings. This analysis of the Gs family leaves out 11 receptors tested for coupling to Gs but not to Golf, and Golf couplings are only counted if there is a supported Gs coupling. All source data are available in tab ‘Fig_4’ in Source data 5.
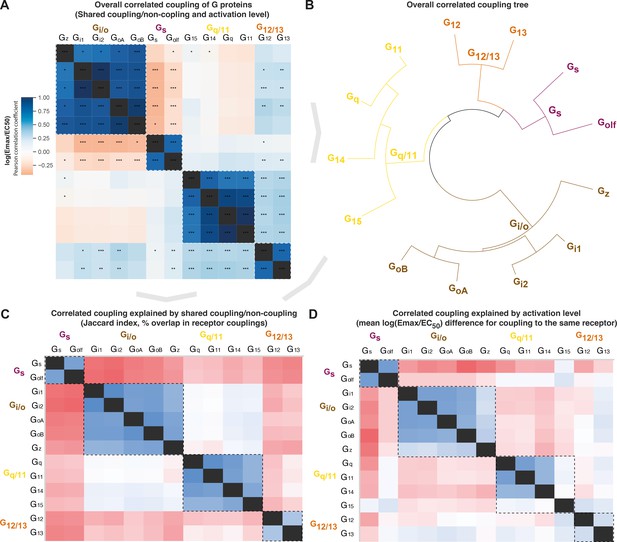
Correlated coupling of G proteins based on their receptor profiles.
(A) Overall correlated coupling of G proteins quantified by the Pearson standard correlation coefficient, which gives a measure of the strength of the linear relationship between two G proteins the log(Emax/EC50) value of noncoupling datapoints was set to 0. Statistically significant pairwise correlations are indicated in cells by *p≤0.05; **p≤0.005, and ***p≤0.0005. (B) Overall coupling correlation of G proteins shown as a tree. (C) Correlated coupling explained by shared coupling/noncoupling quantified as Jaccard indices (% of couplings to the same G protein-coupled receptor [GPCRs]). (D) Correlated coupling explained by activation level quantified as the differences in average log(Emax/EC50) when a G protein pair couples to the same receptor. The values are averages of the log(Emax/EC50) averages of Bouvier and Inoue values, except where data is only available in one dataset, that is, Bouvier only: Gi1-Gi2, GoA-GoB, Gq-G11, and Inoue only: Golf-all G proteins. (A, C–D) All G protein couplings are for class A GPCRs and supported by two datasets (Bouvier or Inoue group or Guide to Pharmacology), and their source values are available in tab ‘Fig_5’ in Source data 5. (A–D) For G proteins (all but Golf) and receptors tested by both the Bouvier and Inoue groups, an average of averages from the two groups is used. The GoA and GoB couplings from Bouvier were compared to the Go values from Inoue which does not distinguish isoforms.
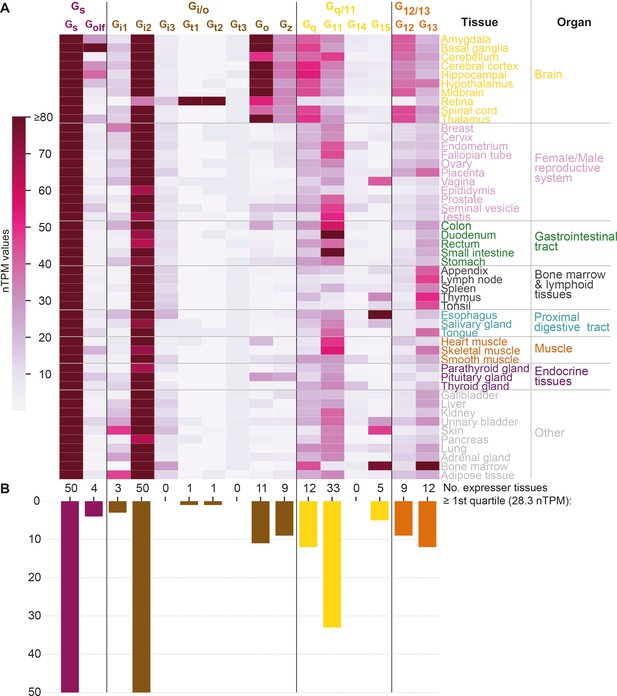
G protein tissue and organ expression profiles.
(A) Expression heatmap of all 16 human G proteins across 50 tissues and 16 organs (here grouped in 8 categories) extracted from the Human Protein Atlas (Uhlén et al., 2015), which also includes data from the Genotype-Tissue Expression project (Lonsdale et al., 2013). The coloring denotes the normalized transcripts per million (nTPM) for each G protein and tissue capped at the median of all maximum G protein expression values (Gt3 at 80 nTPM in retina). (B) Number of tissues for which the given G protein has an expression ≥ the first quartile expression threshold (28.4 nTPM) across all 16 G proteins and 50 tissues.
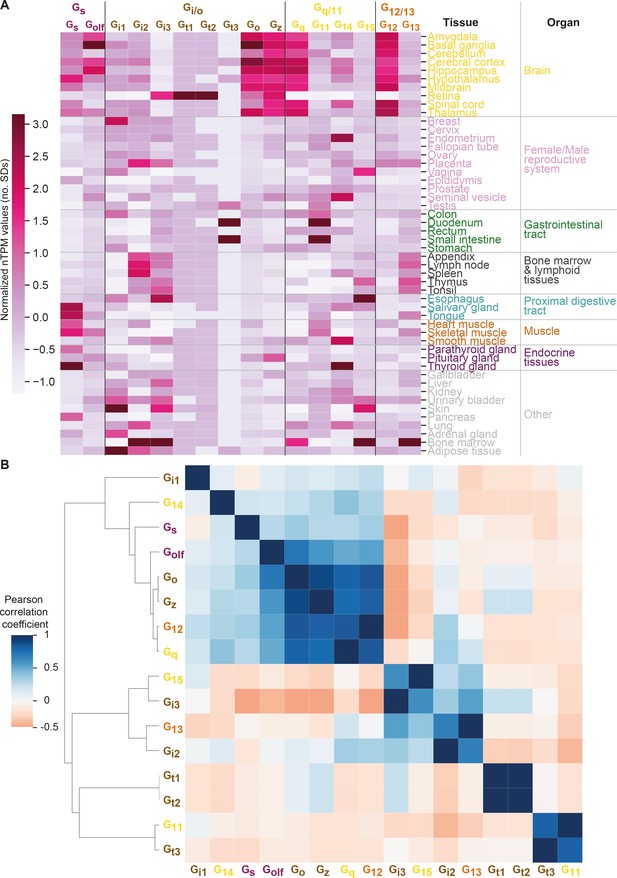
Correlated expression of G proteins.
(A) Expression heatmap like in Figure 6 but with a z-transformation across tissues, thereby visualizing their relative expression for each G protein. (B) Correlated G protein expression across tissues as a function of normalized transcripts per million (nTPM), which is a consensus normalized transcript expression value across the Genotype-Tissue Expression and Human Protein Atlas datasets (see Materials and methods). Pairwise correlation of nTPM expression values by Pearson standard correlation coefficient gives positive and negative correlations between G protein pairs. Expression clusters are distinct from the G protein families, which are denoted with color-coded labels.
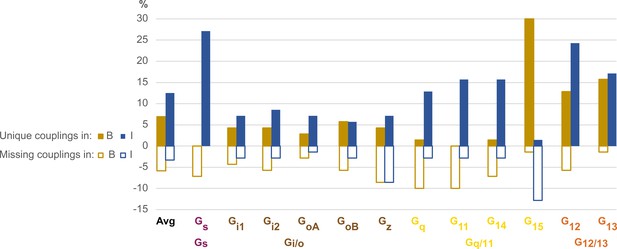
Unique and missing couplings in the Bouvier and Inoue datasets.
Unique and missing couplings (defined in Table 2) among all 70 receptors common to Bouvier, Inoue, and Guide to Pharmacology (GtP) (the 100% includes noncoupling receptors). Missing couplings are shown as negative values. For GtP, we consider coupling to a G protein subtype possible if a coupling has been observed for the respective family. Unique couplings are hidden by default in the online G protein couplings browser in GproteinDb, as they await the independent support by a second group (Pándy-Szekeres et al., 2022).
Tables
G proteins tested by Bouvier et al. and Inoue et al.
The two published datasets contain 11 common G proteins (Gs, Gi1, Gi2, Go, Gz, Gq, G11, G14, G15, G12, and G13), two Inoue et al. specific G proteins (Golf and Gi3) and two Bouvier specific isoforms (GoA and GoB) of the Go protein that spring from the same gene (GNAO1). Inoue et al. Gq chimera replacing the six C-terminal amino acids have identical sequences for Gi1-2, GoA-B, and Gq and G11. All analyses herein used the 11 common G proteins, a Go average of the GoA and GoB isoforms and left out Golf and Gi3 (not present in Bouvier et al.) while identical chimeras from Inoue et al. were used to represent the both members of the pairs Gi1-2, GoA-B, and Gq and G11, respectively. Abbreviations: wt: wildtype; RlucII: Renilla luciferase 2.
| Family | G protein(protein) | Gene name | UniProt name | UniProt identifier | Bouvier | Inoue |
|---|---|---|---|---|---|---|
| Gs | Gs | GNAS | GNAS2 | P63092 | wt +RlucII at pos 67 | Gq1-353-RQYELL |
| Golf | GNAL | GNAL | P38405 | - | Gq1-353-KQYELL | |
| Gi/o | Gi1 | GNAI1 | GNAI1 | P63096-1 | wt | Gq1-353- KDCGLF |
| Gi2 | GNAI2 | GNAI2 | P04899-1 | wt | ||
| Gi3 | GNAI3 | GNAI3 | P08754 | - | Gq1-353-KDCGLY | |
| GoA | GNAO1 | GNAO | P09471-1 | wt | Gq1-353-RGCGLY | |
| GoB | GNAO1 | GNAO | P09471-2 | wt | ||
| Gz | GNAZ | GNAZ | P19086-1 | wt | Gq1-353-KYIGLC | |
| Gt1 (transducin) | GNAT1 | GNAT1 | P11488 | - | Identical toGq1-353-Gi1-2chimera | |
| Gt2 (transducin) | GNAT2 | GNAT2 | P19087 | - | ||
| Ggust (gustducin) | GNAT3 | GNAT3 | A8MTJ3 | - | ||
| Gq/11 | Gq | GNAQ | GNAQ | P50148-1 | wt | wtGq1-353-KEYNLV |
| G11 | GNA11 | GNA11 | P29992-1 | wt | ||
| G14 | GNA14 | GNA14 | O95837-1 | wt | Gq1-353-REFNLV | |
| G15 | GNA15 | GNA15 | P30679-1 | wt | Gq1-353-DEINLL | |
| G12/13 | G12 | GNA12 | GNA12 | Q03113-1 | wt | Gq1-353-KDIMLQ |
| G13 | GNA13 | GNA13 | Q14344-1 | wt | Gq1-353- KQLMLQ |
Terms and definitions used to classify G protein-coupled receptor (GPCR-G) protein couplings.
| Term | Definition |
|---|---|
| Supported | Coupling or noncoupling datapoints that are supported by at least one other dataset, that is, at least two in total. An exception is made for couplings reported only in the GtP database, and not yet tested in a quantitative dataset, as the couplings in GtP are in most cases supported by multiple independent publications. |
| Novel | Coupling that is supported by Bouvier and Inoue but not present in GtP. |
| Proposed | Coupling identified in one but not yet tested in a second quantitative dataset (here Bouvier or Inoue). This refers to mainly receptors, but also G protein subtypes, that have only been tested in one quantitative dataset. Couplings reported only in the GtP database are not considered ‘proposed’ but ‘supported’ because they in most cases are based on the annotation of multiple independent publications. |
| Unique | Coupling that is unique in one dataset (other datasets have no coupling). |
| Missing | Noncoupling in the given dataset but coupling in the other compared datasets. |
-
GtP: Guide to Pharmacology.
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/74107/elife-74107-transrepform1-v2.docx
-
Source data 1
SD cut-off and pEC50 and Emax distributions.
This spreadsheet contains a calculation of a standard deviation (SD) cut-off that gives the best agreement between G protein couplings from the Bouvier and Inoue groups. The cut-off, 1.4 represent the number of standard deviations of a G protein coupling average (at least from triplicates) from the basal signal of the given receptor. The spreadsheet also contains a DataStats tab detailing the distribution of raw and normalized pEC50 and Emax values.
- https://cdn.elifesciences.org/articles/74107/elife-74107-data1-v2.xlsx
-
Source data 2
Assessment of effect of different ligands.
This spreadsheet contains a comparison of the relative average difference obtained for G protein couplings when determined with and without using the same ligand in the Bouvier and Inoue groups.
- https://cdn.elifesciences.org/articles/74107/elife-74107-data2-v2.xlsx
-
Source data 3
Qualitative coupling comparisons.
This spreadsheet contains a qualitative comparison (coupling and non-coupling) of GPCR-G protein pairs from the Bouvier and Inoue groups, and from the Guide to Pharmacology database.
- https://cdn.elifesciences.org/articles/74107/elife-74107-data3-v2.xlsx
-
Source data 4
Normalization and aggregation into G protein families.
This spreadsheet contains a comparative analysis of which normalization and aggregation into G protein families that gives the best agreement between G protein couplings from the Bouvier and Inoue groups.
- https://cdn.elifesciences.org/articles/74107/elife-74107-data4-v2.xlsx
-
Source data 5
Unified coupling map.
This spreadsheet contains a unification of G protein couplings from the Bouvier and Inoue groups, and from the Guide to Pharmacology database into a common coupling map.
- https://cdn.elifesciences.org/articles/74107/elife-74107-data5-v2.xlsx





