Centrally expressed Cav3.2 T-type calcium channel is critical for the initiation and maintenance of neuropathic pain
Figures
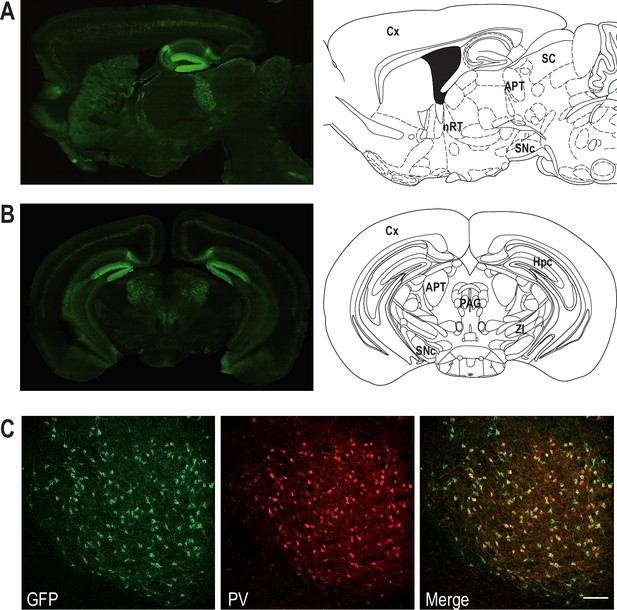
Coexpression of Cav3.2 and parvalbumin (PV) in APT neurons.
Left panels: Cav3.2-GFP immunostaining on a parasagittal (A) and a coronal (B) section of KI mice brains. Right panels: corresponding Mouse Brain Atlas slides from Paxinos and Franklin (parasagittal: 1.08 mm lateral from Bregma; coronal: 2.8 mm posterior from Bregma). APT: anterior pretectum; Cx: cortex; Hpc: hippocampus; nRT: nucleus reticularis thalami; PAG: periaqueductal grey; SC: superior colliculi; SNc: substantia nigra pars compacta; ZI: zona incerta. (C) Confocal microscopy images of a coronal KI mouse brain section of the APT with GFP (green) and PV (red) co-labeling. Scale bar: 100 µm.
-
Figure 1—source data 1
Quantification of Cav3.2-GFP- and parvalbumin (PV)-expressing neurons.
- https://cdn.elifesciences.org/articles/79018/elife-79018-fig1-data1-v2.xlsx
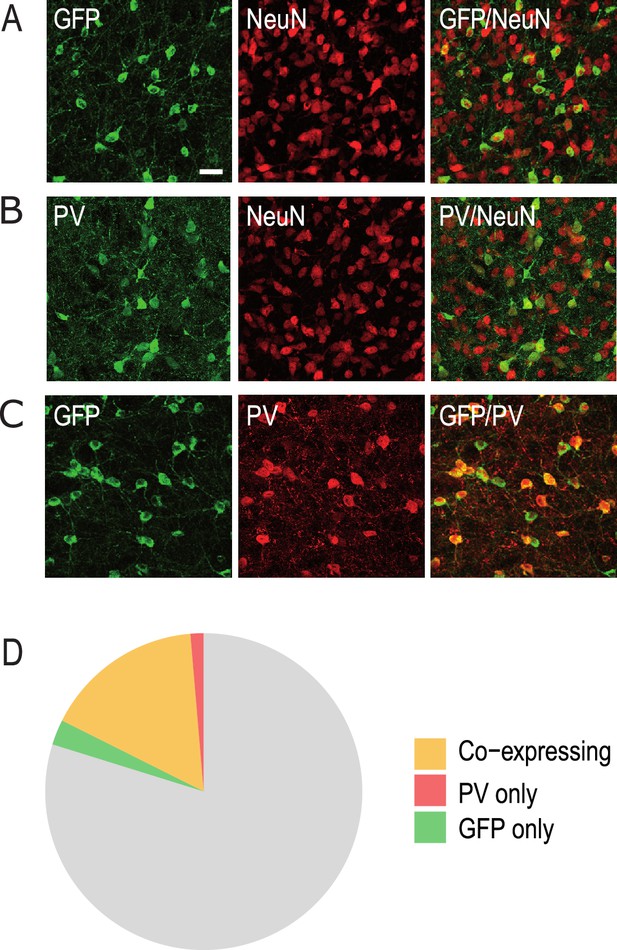
Anterior pretectum (APT) neurons expressing GFP and parvalbumin (PV).
Confocal microscopy images of the co-labelings performed in the APT to estimate the proportion of GFP neurons (A: GFP in green, NeuN in red) and PV neurons (B: PV in green, NeuN in red) and the overlap between these populations (C: GFP in green, PV in red). Scale bar: 30 µm. (D) Diagram representing the percentage of neurons expressing GFP and PV in the APT.
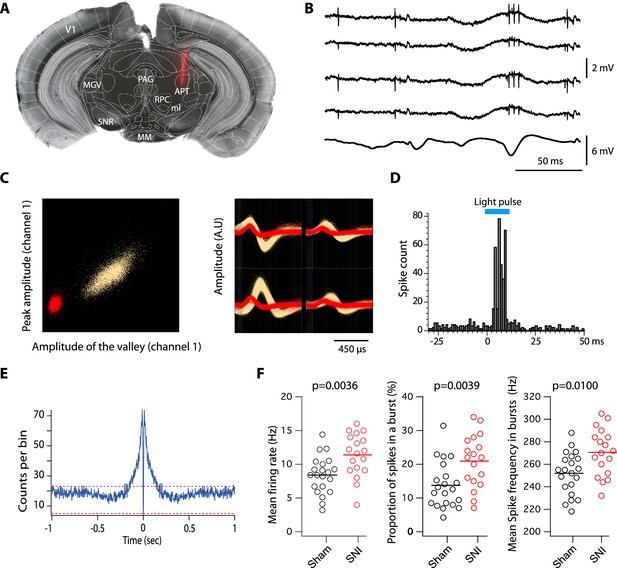
Impact of spared nerve injury (SNI) in PV+ anterior pretectum (APT) neurons.
(A) Example image of a DiI track left by a recording electrode inserted into the APT. Red: DiI. (B) Raw signals from the four wires of a tetrode. Bottom trace shows the simultaneous EEG recording. (C) Left panel: example of a tetrode recording where two units were isolated. All detected action potentials are plotted as their waveform’s amplitude from channel 1 versus the amplitude of their waveform’s valley from channel 1 (in arbitrary units, A.U.). Using this feature space arrangement, two single-unit clusters were isolated (in red and yellow). Right panel: superimposed color-coded action potential waveforms captured by each recording site of the tetrode are shown for the two identified single units (the polarity of the signals is inverted). (D) Example of peristimulus time histograms illustrating spiking response to optogenetic stimulation (10 ms long, represented in blue) over 100 trials of a unit categorized into the PV+ category. (E) Autocorrelogram of recorded single unit for one example cell. 1 ms bins were used. Red dotted lines represent 99% confidence intervals. (F) Scatter dot plots of mean firing rate (left panel), proportion of spikes within a burst (middle panel), and mean spike frequency within a burst (right panel) for fast-bursting APT cells recorded in sham (n = 21 cells, 5 animals) and SNI (n = 18 cells, 4 animals) mice. p values for statistical comparisons were obtained using Wilcoxon sum rank test.
-
Figure 2—source data 1
Firing properties of parvalbumin (PV)-expressing neurons after spared nerve injury (SNI).
- https://cdn.elifesciences.org/articles/79018/elife-79018-fig2-data1-v2.xlsx
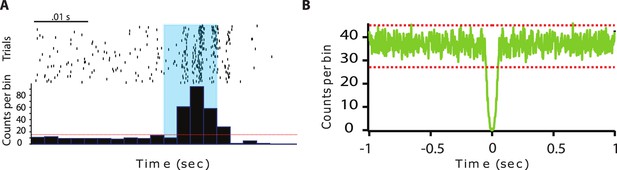
Anterior pretectum (APT) regular spiking neurons.
(A) Example of peristimulus time histograms illustrating spiking response to optogenetic stimulation (10 ms long, represented in blue) over 100 trials of a unit categorized into the PV+ category. (B) Autocorrelogram of recorded single unit for one example cells. 1 ms bins were used. Red dotted lines represent 99% confidence intervals.
-
Figure 2—figure supplement 1—source data 1
Firing properties of PV+ regular spiking neurons.
- https://cdn.elifesciences.org/articles/79018/elife-79018-fig2-figsupp1-data1-v2.xlsx
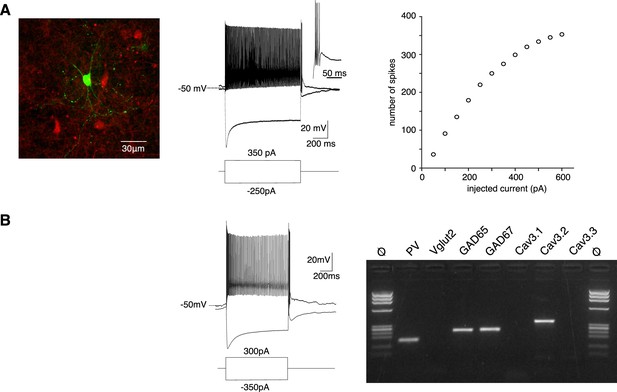
PV+ neurons expressing Cav3.2 channels are GABAergic fast-spiking neurons.
(A) Left image: recorded neuron filled with biocytin (green). Neurons in red are nonrecorded PV+ neurons. Middle traces: depolarizing current injection evokes characteristic fast-spiking activity. Hyperpolarizing current injection evoked a pronounced sag and a rebound high-firing burst upon repolarization. Inset: enlargement of the bursting activity. Right graph: number of spikes evoked by 1 s long depolarizing current injection of increasing amplitudes. (B) Typical example of activities and scRT-PCR products observed in a PV+ neuron. Note the expression of the Cav3.2 channel in the GAD65- and 67-positive neuron.
-
Figure 3—source data 1
Excitability properties of PV+ neurons.
- https://cdn.elifesciences.org/articles/79018/elife-79018-fig3-data1-v2.xlsx
-
Figure 3—source data 2
Original file of the full raw unedited gel shown in Figure 3B.
- https://cdn.elifesciences.org/articles/79018/elife-79018-fig3-data2-v2.zip
-
Figure 3—source data 3
Molecular profile of PV+ neurons.
- https://cdn.elifesciences.org/articles/79018/elife-79018-fig3-data3-v2.xlsx
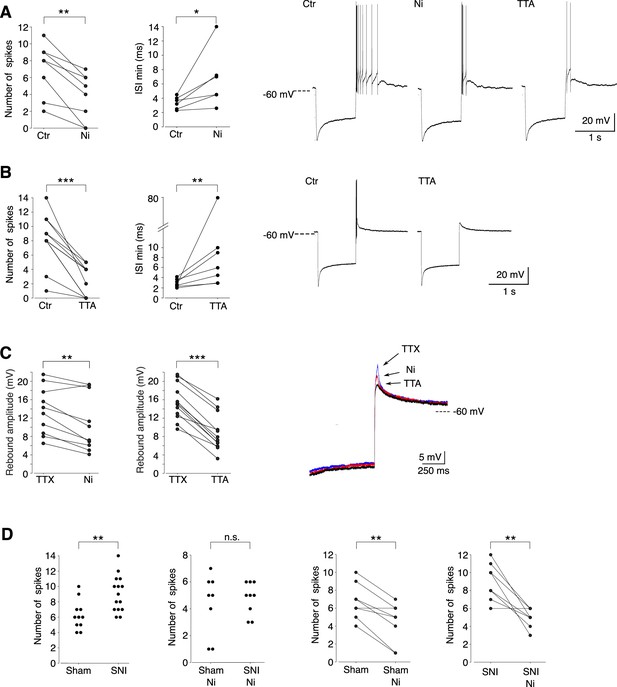
Cav3.2 channel contribution to the bursting activity of PV+ neurons is enhanced in spared nerve injury (SNI) mice.
(A) Effect of 100 µM Ni2+ applications on the number of spikes (left graph, n = 8) and the minimal interspike intervals (ISIs; right graph, n = 6) of the rebound bursts. Typical examples of these pharmacological effects are shown on the right. (B) Effect of 1 µM TTAP2 applications on the number of spikes (left graph, n = 11) and the minimal ISIs (right graph, n = 7) of the rebound bursts. Typical examples of these pharmacological effects are shown on the right. (C) Effect of 100 µM Ni2+ (left graph, n = 10) and 1 µM TTAP2 (right graph, n = 12) applications on the amplitude of the rebound depolarization observed in the presence of 0.5 µM TTX. A typical example is presented in superimposed traces presented on the right. (D) The two left graphs compare the maximal number of spikes of the rebound bursts evoked in neurons of sham-operated and SNI mice in control condition (n = 12 and 15, respectively) and after application of 100 µM Ni2+ (n = 8 and 9, respectively). The effects of Ni2+ application on each neuron are presented in the two right graphs (n = 8 and 9 for sham and SNI, respectively). A, B, C, D right graphs: Wilcoxon signed rank test; D left graphs: Wilcoxon sum rank test. ***p < 0.001 ; **p: 0.001 < p < 0.01 ; *0.01 < p < 0.05.
-
Figure 4—source data 1
Ni and TTA effects on burst properties of PV+ neurons.
- https://cdn.elifesciences.org/articles/79018/elife-79018-fig4-data1-v2.xlsx
-
Figure 4—source data 2
Ni and TTA effects on the rebound depolarization in PV+ neurons.
- https://cdn.elifesciences.org/articles/79018/elife-79018-fig4-data2-v2.xlsx
-
Figure 4—source data 3
Burst properties of PV+ neurons in slices from sham and spared nerve injury (SNI) mice.
- https://cdn.elifesciences.org/articles/79018/elife-79018-fig4-data3-v2.xlsx
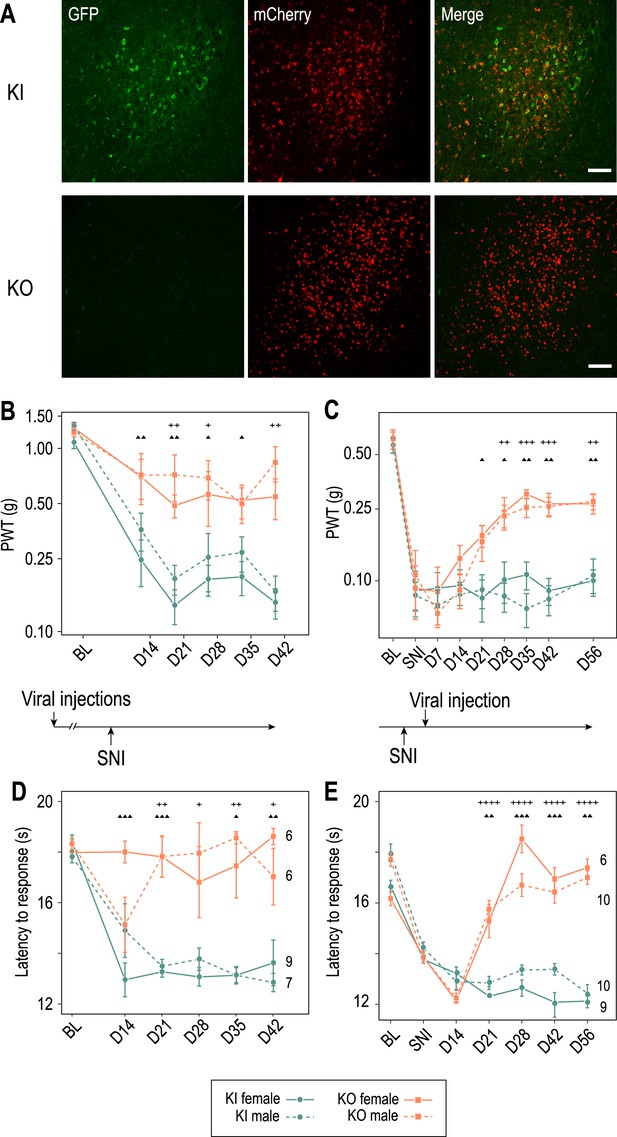
Cav3.2 preventive and therapeutic knockout in the anterior pretectum (APT) alleviates neuropathy induced by spared nerve injury (SNI).
(A) Confocal microscopy images of GFP (green) and mCherry (red) co-labeling in the APT of KI-Cav3.2-GFP (Control) mice and KO-Cav3.2-APT (KO) bilaterally injected in the APT with AAV8-hSyn-mCherry and AAV8-hSyn-mCherry-CRE virus, respectively, and further tested for mechanical and cold sensitivity. Scale bar: 100 µm. Neuropathic behaviors were tested in male (dashed lines) and female (solid lines) mice with preventive (B, D) and therapeutic (C, E) KO of Cav3.2 in the APT (orange, ■), and in control KI mice (green, ●). (B, C) Mechanical sensitivity was assessed by measuring paw withdrawal thresholds (PWTs) in response to Von Frey filaments stimulations using the up-and-down method. (D, E) Cold sensitivity was assessed by measuring the paw withdrawal latency in response to immersion in 18°C water. For preventive KO (B, D) mice were tested prior to the SNI (BL) and once a week during the 6 subsequent weeks (days 14–48). For therapeutic KO (C, E) mechanical and cold sensitivity were assessed before (BL) and 14 days after SNI (SNI), and tested during several weeks following the subsequent viral injections (days 14–56 after viral injection). (B–E) Wilcoxon sum rank test for female KO versus KI and male KO versus KI comparison at each time point. ++++ : p < 0.0001, +++ or ▲▲▲: 0.0001 < p < 0.001, ++ or ▲▲: 0.001 < p < 0.01, + or ▲: 0.01 < p < 0.05 (males: +; females: ▲).
-
Figure 5—source data 1
Characterization of the effect of APT-Cav3.2 preventive or curative knockout on neuropathic pain induced by spared nerve injury (SNI).
- https://cdn.elifesciences.org/articles/79018/elife-79018-fig5-data1-v2.xlsx
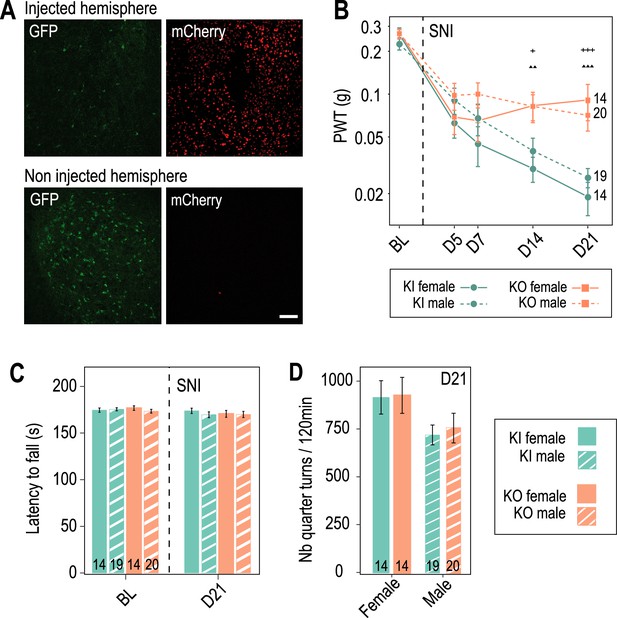
Preventive Cav3.2 knockout in the anterior pretectum (APT) has no impact on motor coordination and spontaneous locomotion.
(A) Confocal microscopy images of GFP labeling (green) and mCherry expression (red) in the APT of a KI mouse unilaterally injected with AAV8-hSyn-Cre-mCherry virus (up panel: injected hemisphere; down panel: noninjected hemisphere). Scale bar: 100 µm. Note the drastic reduction in the number of GFP+ neurons observed 2 weeks post viral injection in the injected hemisphere. (B–D) Motor coordination and spontaneous locomotion assessed in control KI mice (green, ●) and mice with preventive APT-Cav3.2 KO (orange, ■) subjected to spared nerve injury (SNI). (B) Control of the development of mechanical allodynia in male (dashed lines) and female (solid lines) mice. Note that as for the cohorts presented in Figure 5, preventive Cav3.2 KO reduced allodynia. Wilcoxon sum rank test for female KO versus KI and male KO versus KI comparison at each time point. +++ or ▲▲▲: 0.0001 < p < 0.001, ▲▲: 0.001 < p < 0.01, +: 0.01 < p < 0.05 (males: +; females: ▲). (C) Latency to fall from an accelerating rotarod wheel (0–16 rpm over 3 min) was measured in males (dashed bars) and females (solid bars), before (BL) and after (day 21) SNI surgery. No statistical differences were detected between groups and before and after SNI surgery within a group using Wilcoxon sum rank test and Wilcoxon signed rank test, respectively. (D) The number of quarter turns was detected in a circular corridor over 120 min, measured in males (dashed bars) and females (solid bars), 21 days after SNI surgery. No significant differences were detected between groups using the Wilcoxon sum rank test.
-
Figure 5—figure supplement 1—source data 1
Characterization of preventive APT-Cav3.2 knockout effect on mechanical sensitivity and locomotion.
- https://cdn.elifesciences.org/articles/79018/elife-79018-fig5-figsupp1-data1-v2.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus, male and female) | Cav3.2-eGFPflox | PMID:25600872 (François et al., 2015) | Cacna1htm1.1(epH)Ebo/J | KI mice with two loxP site-flanked ecliptic-GFP tag into exon 6 of the Cacna1h gene |
| Strain, strain background (Mus musculus, female) | PV-Cre | Jackson lab | B6.129P2- Pvalb-tm1(cre)Arbr/J | Cre recombinase expression in Pvalb-expressing cells |
| Strain, strain background (Mus musculus, male) | Ai14 | Jackson lab | B6.Cg-Gt(ROSA) 26Sor-tm14(CAG-tdTomato)Hze/J | Cre-dependent expression of the red fluorescent protein variant (tdTomato) |
| Strain, strain background (Mus musculus, male) | Ai32 | Jackson lab | B6.Cg-Gt(ROSA) 26Sortm32(CAG-COP4*H134R/EYFP)Hze/J | Cre-dependent expression of the ChR2(H134R)-EYFP |
| Sequence-based reagent | Mouse Pvalb external sense | PMID:21795545 (Tricoire et al., 2011) | PCR primers | GCCTGAAGAAAAAGAACCCG |
| Sequence-based reagent | Mouse Pvalb external antisense | PMID:21795545 (Tricoire et al., 2011) | PCR primers | AATCTTGCCGTCCCCATCCT |
| Sequence-based reagent | Mouse Pvalb internal sense | PMID:21795545 (Tricoire et al., 2011) | PCR primers | CGGATGAGGTGAAGAAGGTGT |
| Sequence-based reagent | Mouse Pvalb internal antisense | PMID:21795545 (Tricoire et al., 2011) | PCR primers | TCCCCATCCTTGTCTCCAGC |
| Sequence-based reagent | Mouse Gad2 external sense | PMID:34766906 (Karagiannis et al., 2021) | PCR primers | CCAAAAGTTCACGGGCGG |
| Sequence-based reagent | Mouse Gad2 external antisense | PMID:34766906 (Karagiannis et al., 2021) | PCR primers | GTGAGCAGTATCGCAGCCCC |
| Sequence-based reagent | Mouse Gad2 internal sense | PMID:34766906 (Karagiannis et al., 2021) | PCR primers | CACCTGCGACCAAAAACCCT |
| Sequence-based reagent | Mouse Gad2 internal antisense | PMID:12196560 (Férézou et al., 2002) | PCR primers | GATTTTGCGGTTGGTCTGCC |
| Sequence-based reagent | Mouse Gad1 external sense | PMID:23565079 (Cabezas et al., 2013) | PCR primers | TACGGGGTTCGCA CAGGTC CGGGCGG |
| Sequence-based reagent | Mouse Gad1 external antisense | PMID:23565079 (Cabezas et al., 2013) | PCR primers | CCCAGGCAGCATCCACAT |
| Sequence-based reagent | Mouse Gad1 internal sense | PMID:23565079 (Cabezas et al., 2013) | PCR primers | CCCAGAAGTGAAGACAAAAGGC |
| Sequence-based reagent | Mouse Gad1 internal antisense | PMID:23565079 (Cabezas et al., 2013) | PCR primers | AATGCTCCGTAAACAGTCGTGC |
| Sequence-based reagent | Mouse Slc17a6 external sense | This paper | PCR primers | TGGAGAAGAAGCAGGACAACC |
| Sequence-based reagent | Mouse Slc17a6 external antisense | This paper | PCR primers | GTGAGCAGTATCGCAGCCCC |
| Sequence-based reagent | Mouse Slc17a6 internal sense | This paper | PCR primers | TGACAGAGGACGGTAAGCCCC |
| Sequence-based reagent | Mouse Slc17a6 internal antisense | This paper | PCR primers | TCATCCCCACGGTCTCGG |
| Sequence-based reagent | Mouse Cacna1g external sense | This paper | PCR primers | CACCGATGTCACTGCCCAAG |
| Sequence-based reagent | Mouse Cacna1g external antisense | This paper | PCR primers | GGCTCTCCTGACCCTCTCCA |
| Sequence-based reagent | Mouse Cacna1g internal sense | This paper | PCR primers | GCTCTCGCCGCACCAGTA |
| Sequence-based reagent | Mouse Cacna1g internal antisense | This paper | PCR primers | CTTGGGCTCCTACGCTTCAG |
| Sequence-based reagent | Mouse Cacna1h external sense | This paper | PCR primers | TACCAGACAGAGGAGGGCGA |
| Sequence-based reagent | Mouse Cacna1h external antisense | This paper | PCR primers | CTATCACCACCAGGCACAGG |
| Sequence-based reagent | Mouse Cacna1h internal sense | This paper | PCR primers | CATTCATCTGCTCCTCACGC |
| Sequence-based reagent | Mouse Cacna1h internal antisense | This paper | PCR primers | GCCCACAATGATGAGGAGGA |
| Sequence-based reagent | Mouse Cacna1i external sense | This paper | PCR primers | GTCCCCCTCCATCCCCTC |
| Sequence-based reagent | Mouse Cacna1i external antisense | This paper | PCR primers | CAATGAAGAAGTCCAAGCGGTT |
| Sequence-based reagent | Mouse Cacna1i internal sense | This paper | PCR primers | GTTGCCTTCTTCTGCCTGCG |
| Sequence-based reagent | Mouse Cacna1i internal antisense | This paper | PCR primers | TCCCCGAGGTAGCACTTCTT |
| Strain, strain background (AAV) | AAV8-hSyn-mCherry-Cre | UNC Vector Core | ||
| Strain, strain background (AAV) | AAV8-hSyn-mCherry | UNC Vector Core | ||
| Antibody | Anti-GFP (chicken polyclonal) | Life Technologies | A10262 | 1:500 |
| Antibody | Anti-GFP (rabit polyclonal) | Chromotek | PABG1 | 1:500 |
| Antibody | Anti-Parvalbumin (mouse monoclonal) | Sigma-Aldrich | P3088 | 1:1000 |
| Antibody | Anti-NeuN (rabbit polyclonal) | Merck Millipore | ABN78 | 1:500 |
| Antibody | Anti-chicken-Alexa Fluor 488 (goat polyclonal) | Life Technologies | A11039 | 1:500 |
| Antibody | Anti-chicken-Alexa Fluor 488 (donkey polyclonal) | Sigma-Aldrich | SAB4600031 | 1:500 |
| Antibody | Anti-rabit-Alexa Fluor 647 (goat polyclonal) | Life Technologies | A21244 | 1:500 |
| Antibody | Anti-mouse-Alexa Fluor 488 (goat polyclonal) | Life Technologies | A11001 | 1:500 |
| Antibody | Anti-rabit-Cyanine Cy3 (donkey polyclonal) | Jackson ImmunoResearch Europe | 711-165-1525 | 1:2000 |
| Chemical compound, drug | Isoflurane | Iso-Vet | Cat# 3248850; GTIN: 18904026625157 | |
| Chemical compound, drug | Buprenorphine (Buprecare) | Axience | GTIN: 03760087151893 | |
| Chemical compound, drug | Pentobarbital (Euthasol Vet) | Dechra | GTIN: 08718469445110 | |
| Chemical compound, drug | Ketamine | Imalgene 1000 | GTIN: 03661103003199 | |
| Chemical compound, drug | Xylazine (Rompun) | Bayer | GTIN: 04007221032311 | |
| Chemical compound, drug | Bupivacaine | Henry Schein | Cat# 054879 | |
| Chemical compound, drug | Twelve TVM Eye Support Drops | TVM UK Animal Health | GTIN: 03700454507502 | |
| Chemical compound, drug | Vetedine | Vetoquinol | GTIN: 03605870001385 | |
| Chemical compound, drug | NaCl | Sigma-Aldrich | Cat# S6191; CAS: 7647-14-5 | |
| Chemical compound, drug | KCl | Sigma-Aldrich | Cat# 60128; CAS: 7447-40-7 | |
| Chemical compound, drug | CaCl2 | Sigma-Aldrich | Cat# C3881; CAS: 10035-04-8 | |
| Chemical compound, drug | MgCl2 | Sigma-Aldrich | Cat# M2670; CAS: 7791-18-6 | |
| Chemical compound, drug | NaH2PO4 | Sigma-Aldrich | Cat# S5011; CAS: 7558-80-7 | |
| Chemical compound, drug | NaHCO3 | Sigma-Aldrich | Cat# 31437; CAS: 144-55-8 | |
| Chemical compound, drug | Glucose | Sigma-Aldrich | Cat# 49159; CAS: 14431-43-7 | |
| Chemical compound, drug | Potassium gluconate | Sigma-Aldrich | Cat# G4500; CAS: 299-27-4 | |
| Chemical compound, drug | EGTA | Sigma-Aldrich | Cat# E3889; CAS: 67-42-5 | |
| Chemical compound, drug | HEPES | Sigma-Aldrich | Cat# H3375; CAS: 7365-45-9 | |
| Chemical compound, drug | Disodium ATP | Sigma-Aldrich | Cat# A7699; CAS: 34369-07-8 | |
| Chemical compound, drug | Biocytin | Sigma-Aldrich | Cat# B4261; CAS: 576-19-2 | |
| Chemical compound, drug | D-Mannitol | Sigma-Aldrich | Cat# M9647; CAS: 69-65-8 | |
| Chemical compound, drug | Sodium Pyruvate | Sigma-Aldrich | Cat# P2256; CAS: 113-24-6 | |
| Chemical compound, drug | Kynurenic acid | Hello Bio | Cat# HB0363; CAS: 2439-02-3 | |
| Chemical compound, drug | Paraformaldehyde in 0.1 M phosphate buffer saline | Electron Microscopy Sciences | Cat# 15710; CAS: 30525-89-4 | |
| Chemical compound, drug | Trizma | Sigma-Aldrich | Cat# T1503; CAS: 77-86-1 | |
| Chemical compound, drug | Sodium azide | Sigma-Aldrich | Cat# S2002; CAS: 26628-22-8 | |
| Chemical compound, drug | Phosphate buffer saline | Sigma-Aldrich | Cat# D1408 | |
| Chemical compound, drug | Triton X-100 | Sigma-Aldrich | Cat# T8787; CAS: 9002-93-1 | |
| Chemical compound, drug | Fluoromount Aquaous Mounting Medium | Sigma-Aldrich | Cat# F4680 | |
| Chemical compound, drug | Donkey serum | Sigma-Aldrich | Cat# D9663 | |
| Chemical compound, drug | Streptavidin- Rhodamine-RedX | Jackson ImmunoResearch Europe | Cat# 016-290-084 | |
| Chemical compound, drug | DiI, 10% in ethanol | Invitrogen | Cat# V22885 | |
| Chemical compound, drug | CNQX | Hello bio | Cat# HB02057; CAS: 479347-85-8 | |
| Chemical compound, drug | SR95531 hydrobromide | Hello bio | Cat# HB0901; CAS: 104104-50-9 | |
| Chemical compound, drug | TTX | Latoxan | Cat# L8503; CAS: 4368-28-9 | |
| Chemical compound, drug | Nickel chloride hydrate | Sigma-Aldrich | Cat# 364304; CAS: 69098-15-3 | |
| Chemical compound, drug | TTA-P2 | Alomone labs | Cat# T-155; CAS: 918430-49-6 | |
| Chemical compound, drug | Taq DNA Polymerase | Qiagen | 201205 | |
| Chemical compound, drug | RNasin Ribonuclease Inhibitors | Promega | N2511 | |
| Chemical compound, drug | SuperScript II Reverse Transcriptase | Invitrogen | Cat# 18064014 | |
| Software, algorithm | Pclamp V 10.2 | Molecular Devices | https://www.moleculardevices.com/ | |
| Software, algorithm | Matlab 2019b | MathWorks | https://fr.mathworks.com/ | |
| Software, algorithm | Igor Pro V6 | Wavemetrics | https://www.wavemetrics.com | |
| Software, algorithm | Cheetah V 5 | Neuralynx | https://neuralynx.com/software/cheetah | |
| Software, algorithm | KlustaKwik | Neuralynx | https://neuralynx.com/software/cheetah | |
| Software, algorithm | SpikeSort3D V 2 | Neuralynx | https://neuralynx.com/software/cheetah | |
| Software, algorithm | Fiji/ImageJ | NIH | https://imagej.nih.gov/ij/download.html | |
| Software, algorithm | Rstudio | https://www.rstudio.com/ | ||
| Software, algorithm | R version 4.1.0 (2021-05-18) | Camp Pontanezen – The R Foundation for Statistical Computing | https://www.r-project.org/ | |
| Other | Quartz-insulated platinum/tungsten (90%/10%) tetrodes | Thomas Recording GmbH | Cat# AN000259 | See ‘In vivo electrophysiological recordings’ in the Method Details |
| Other | Optical fibers | Thomas Recording GmbH | Cat# AN000514 | Optical fiber used to deliver 470-nm blue-light pulse for Photo-assisted Identification of Neuronal Population |
| Other | Borosilicate glass capillaries | Hilgenberg | Cat# 1409250 | See ‘In vitro whole-cell patch-clamp recording’ in the Method Details |
| Other | 4-0 Vicryl | Ethicon | ref JV397 | See ‘Virus stereotaxic injections’ in the Method Details |





