Neuroendocrine gene expression coupling of interoceptive bacterial food cues to foraging behavior of C. elegans
Figures

Ingestion of bacterial food inhibits daf-7 expression in the ASJ neurons.
(A) Schematic of experimental exposure to varied food conditions. Animals were grown to adulthood on edible E. coli OP50, then transferred to various experimental food conditions: ‘Fed’: E. coli OP50; ‘Aztreonam’: OP50 treated with aztreonam; ‘Bottom’: OP50 was seeded underneath the agar of the plate; ‘Lid’: Animals placed on agar with no food but where food was seeded on a spot of agar on the inside of the lid of the plate; ‘Empty’: no food. (B) Model for the convergence of external and ingested food signals on daf-7 expression in the ASJ neurons. (C) pdaf-7::gfp expression pattern in animals under different food conditions, from left to right: ‘Fed,’ ‘Aztreonam,’ ‘Bottom,’ ‘Lid,’ ‘Empty.’ Filled triangles indicate the ASI neurons; open triangles indicate the ASJ neurons. Scale bar indicates 50 µm. (D, E, F), Maximum fluorescence values of pdaf-7::gfp in the ASJ neurons of adult animals under various food conditions. Each point represents an individual animal, and error bars indicate standard deviation. ****p<0.0001, ns, not significant as determined by an unpaired two-tailed t-test. (G) Maximum fluorescence values of pdaf-7::gfp in the ASJ neurons of adult animals at various time points after being moved from ‘Fed’ to ‘Bottom’ conditions. Each point represents an individual animal, and error bars indicate standard deviation. ****p<0.0001, *p<0.05, ns, not significant as determined by an unpaired two-tailed t-test. (H) Maximum fluorescence values of pdaf-7::gfp in the ASJ neurons of adult wild-type and del-3(ok2613); del-7(ok1187) under ‘Fed,’ ‘Bottom,’ and ‘Empty’ conditions. Each point represents an individual animal, and error bars indicate standard deviation. ***p<0.001, ns, not significant as determined by an unpaired two-tailed t-test.

daf-7 expression in the ASJ neurons promotes roaming.
(A) Fraction of time roaming (left), and number of squares entered in exploration assay (right) of wild-type, daf-7(e1372), and daf-7(ok3125) animals. Each point represents an individual animal, and error bars indicate standard deviation. ****p<0.0001, **p<0.01, *p<0.05 as determined by an unpaired two-tailed t-test. (B) Fraction of time roaming of wild-type, daf-7(ok3125), and two independent lines where daf-7 cDNA was expressed under the trx-1 promoter in a daf-7(ok3125) background. Each point represents an individual animal, and error bars indicate standard deviation. **p<0.01, *p<0.05 as determined by an unpaired two-tailed t-test. (C) Fraction of time roaming (left), and number of squares entered in exploration assay (right) of wild-type, daf-7(e1372), daf-7(ok3125), and a floxed daf-7 strain with and without Cre expressed under the trx-1 promoter. Each point represents an individual animal, and error bars indicate standard deviation. **p<0.01, *p<0.05 as determined by an unpaired two-tailed t-test. (D) Model of positive-feedback relationship between food ingestion, daf-7 expression in the ASJ neurons, and roaming. (E) Sample trace files of representative individual wild-type, daf-7(e1372), and daf-7(ok3125) animals. R=roaming, D=dwelling. (F) Duration of dwelling (open circles) and roaming (closed circles) states for wild-type, daf-7(e1372), and daf-7(ok3125) animals. Each point represents a discrete roaming or dwelling period. Error bars indicate standard deviation. ***p<0.001, *p<0.05, ns, not significant as determined by an unpaired two-tailed t-test.
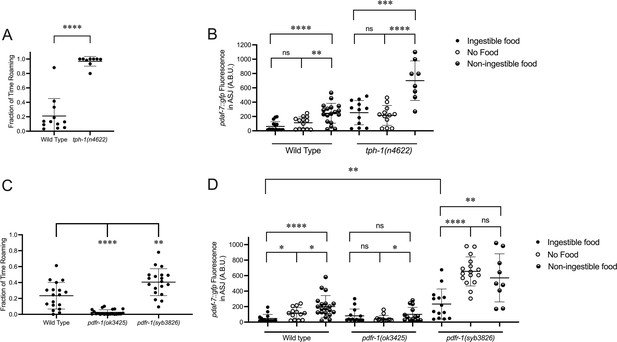
Known modulators of roaming and dwelling affect daf-7 expression in the ASJ neurons.
(A) Fraction of time roaming of wild-type and tph-1(n4622) animals. Each point represents an individual animal, and error bars indicate standard deviation. ****p<0.0001 as determined by an unpaired two-tailed t-test. (B) Maximum fluorescence values of pdaf-7::gfp in the ASJ neurons of adult wild-type and tph-1(n4622) animals under ‘Fed,’ ‘Empty,‘ and ‘Bottom’ conditions. Each point represents an individual animal, and error bars indicate standard deviation. ****p<0.0001, ***p<0.001, **p<0.01, ns, not significant as determined by an unpaired two-tailed t-test. (C) Fraction of time roaming of wild-type, pdfr-1(ok3425), and pdfr-1(syb3826) animals. Each point represents an individual animal, and error bars indicate standard deviation. ****p<0.0001, **p<0.01 as determined by an unpaired two-tailed t-test. (D) Maximum fluorescence values of pdaf-7::gfp in the ASJ neurons of adult wild-type, pdfr-1(ok3425), and pdfr-1(syb3826) animals under ‘Fed,’ ‘Empty,’ and ‘Bottom’ conditions. Each point represents an individual animal, and error bars indicate standard deviation. ****p<0.0001, **p<0.01, *p<0.05, ns, not significant as determined by an unpaired two-tailed t-test.
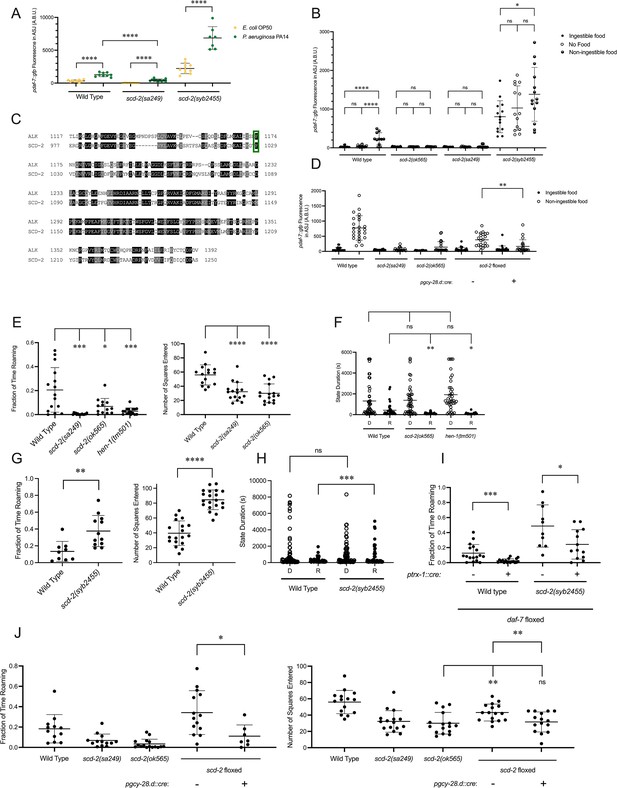
SCD-2 regulates daf-7 expression in the ASJ neurons and roaming behavior.
(A) Maximum fluorescence values of pdaf-7::gfp in the ASJ neurons of adult wild-type, scd-2(sa249), and scd-2(syb2455) animals exposed to E. coli OP50 and P. aeruginosa PA14. Each point represents an individual animal, and error bars indicate standard deviation. ****p<0.0001, as determined by an unpaired two-tailed t-test. (B) Maximum fluorescence values of pdaf-7::gfp in the ASJ neurons of adult wild-type, scd-2(ok565), scd-2(sa249), and scd-2(syb2455) animals under ‘Fed,’ ‘Empty,’ and ‘Bottom’ conditions. Each point represents an individual animal, and error bars indicate standard deviation. ****p<0.0001, *p<0.05, ns, not significant as determined by an unpaired two-tailed t-test. (C) Sequence alignment of the kinase domains of human anaplastic lymphoma kinase (ALK) (top) and C. elegans SCD-2 (bottom). Amino acids highlighted in black are identical and those in gray are similar. F1174/F1029 is outlined in green. (D) Maximum fluorescence values of pdaf-7::gfp in the ASJ neurons of adult wild-type, scd-2(sa249), scd-2(ok565), and floxed scd-2 animals without and with a transgene expressing Cre under the AIA-specific gcy-28.d promoter under ‘Fed’ and ‘Bottom’ conditions. Each point represents an individual animal, and error bars indicate standard deviation. **p<0.01 as determined by an unpaired two-tailed t-test. (E) Fraction of time spent roaming (left) and number of squares entered in exploration assay (right) for wild-type, scd-2(sa249), scd-2(ok565), and hen-1(tm501) animals. Each point represents an individual animal. Error bars indicate standard deviation. ****p<0.0001, ***p<0.001, *p<0.05 as determined by an unpaired two-tailed t-test. (F) Duration of roaming (closed circles) and dwelling (open circles) states for wild-type, scd-2(ok565), and hen-1(tm501). Each point represents a discrete roaming or dwelling period. Error bars indicate standard deviation. **p<0.01, *p<0.05, ns, not significant as determined by an unpaired two-tailed t-test. (G) Fraction of time spent roaming (left) and number of squares entered in exploration assay (right) for wild-type and scd-2(syb2455) animals. Each point represents an individual animal. Error bars indicate standard deviation. ****p<0.0001, **p<0.01 as determined by an unpaired two-tailed t-test. (H) Duration of roaming (closed circles) and dwelling (open circles) states for wild-type and scd-2(syb2455) animals. Each point represents a discrete roaming or dwelling period. Error bars indicate standard deviation. ***p<0.001, ns, not significant as determined by an unpaired two-tailed t-test. (I) Fraction of time roaming for wild-type of scd-2(syb2455) animals with a floxed allele of daf-7 without or with an ASJ-specific cre transgene. ***p<0.001, *p<0.05 as determined by an unpaired two-tailed t-test. (J) Left: Fraction of time spent roaming in worm tracker assay for scd-2(sa249), scd-2(ok565), and floxed scd-2 without or with a transgene expressing Cre under the AIA-specific gcy-28.d promoter. Right: number of squares entered in exploration assay for wild-type, scd-2(sa249), scd-2(ok565), and floxed scd-2 with or without a transgene expressing Cre under the AIA-specific gcy-28.d promoter. Each point represents an individual animal. Error bars indicate standard deviation. **p<0.01, *p<0.05, ns, not significant as determined by an unpaired two-tailed t-test.

DAF-7 expression in the ASJ neurons correlates with an increase in roaming behavior under various conditions.
(A) Maximum fluorescence values of pdaf-7::gfp in the ASJ neurons of adult hermaphrodites and males under ‘Fed’ and ‘Aztreonam’ conditions. Each point represents an individual animal, and error bars indicate standard deviation. **p<0.01 as determined by an unpaired t-test. (B) Fraction of time roaming of wild-type hermaphrodites and males. Each point represents an individual animal, and error bars indicate standard deviation. ****p<0.0001 as determined by an unpaired t-test. (C) Maximum fluorescence values of pdaf-7::gfp in the ASJ neurons of adult hermaphrodites fed E. coli OP50, P. aeruginosa PA14, or P. aeruginosa PA14 ∆gacA. Each point represents an individual animal, and error bars indicate standard deviation. ****p<0.0001, **p<0.01 as determined by an unpaired t-test. (D) Fraction of time roaming of wild-type hermaphrodites on E. coli OP50, P. aeruginosa PA14, or P. aeruginosa PA14 ∆gacA. Each point represents an individual animal, and error bars indicate standard deviation. ****p<0.0001, ***p<0.001, *p<0.05 as determined by an unpaired t-test. (E) daf-7 expression in the ASJ neurons responds to environmental conditions and is correlated with internal state. Under favorable conditions, daf-7 is not expressed in the ASJ neurons, and this is correlated with an internal state that favors exploitation of the animal’s current environment. Under unfavorable conditions, daf-7 expression is induced in the ASJ neurons, consistent with an internal state that favors exploration.
Tables
Complete list of C. elegans strains used in this study.
| Strain Name | Genotype | Source |
|---|---|---|
| N2 | Wild type | Caenorhabditis Genetics Center (CGC) |
| JT249 | scd-2(sa249) | CGC |
| RB783 | scd-2(ok565) | CGC |
| JC2154 | hen-1(tm501) | CGC |
| PHX2455 | scd-2(syb2455) | This study/SunyBiotech |
| FK181 | ksIs2[pdaf-7::gfp; rol-6(su1006)] | CGC |
| ZD2540 | ksIs2; scd-2(sa249) | This study |
| ZD930 | ksIs2; scd-2(ok565) | This study |
| ZD918 | ksIs2; hen-1(tm501) | This study |
| ZD2605 | ksIs2; scd-2(syb2455) | This study |
| CB1372 | daf-7(e1372) | CGC |
| ZD715 | daf-7(ok3125) | Meisel et al., 2014 |
| ZD695 | daf-7(ok3125);qdEx34[ptrx-1::daf-7;pges-1::GFP] | Meisel et al., 2014 |
| ZD696 | daf-7(ok3125);qdEx35[ptrx-1::daf-7;pges-1::GFP] | Meisel et al., 2014 |
| ZD2632 | ksIs2; del-3(ok2613);del-7(ok1187) | This study |
| MT14984 | tph-1(n4622) | Horvitz Lab |
| ZD667 | ksIs2; tph-1(n4622) | This study |
| ZD2079 | pdfr-1(ok3425) | Hilbert and Kim, 2018 |
| PHX3826 | pdfr-1(syb3826) | This study/SunyBiotech |
| ZD1987 | ksIs2; pdfr-1(ok3425); him-5(e1490) | Hilbert and Kim, 2018 |
| ZD2633 | ksIs2; pdfr-1(syb3826); him-5(e1490) | This study |
| ZD2721 | scd-2(syb5845 syb6052) Backcrossed x2 | This study |
| ZD2722 | daf-7(syb5855 syb5965) qdEx[ptrx-1::Cre; pofm-1::gfp] Backcrossed x2 | This study |
| ZD2752 | scd-2(syb5845 syb6052); ex[gcy-28.dp::cre; ofm-1p::gfp] | This study |
| ZD2766 | scd-2(syb5845 syb6052); ex[gcy-28.dp::cre; ofm-1p::gfp]; ksIs2 | This study |
| ZD2796 | scd-2(syb2455); daf-7(syb5855 syb5965) qdEx[ptrx-1::Cre; pofm-1::gfp] | This study |
| ZD1005 | him-5(e1390); ksIs2 | Hilbert and Kim, 2018 |





