Pharmacologically inducing regenerative cardiac cells by small molecule drugs
Figures
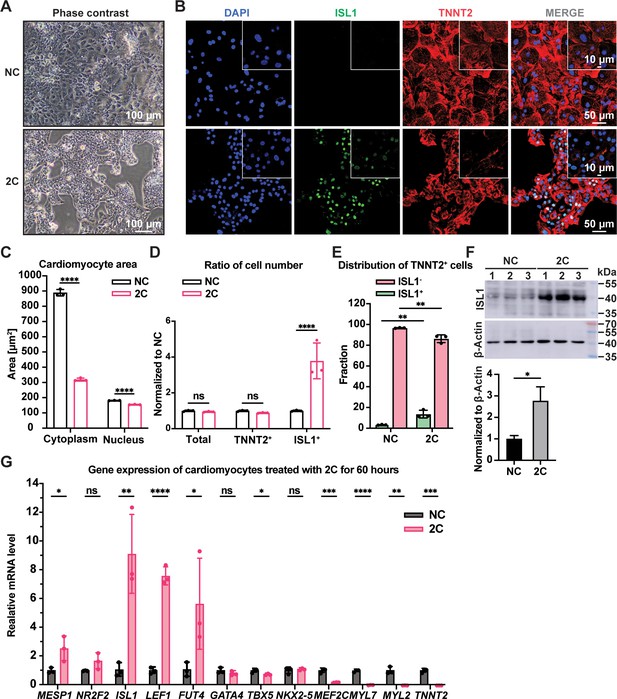
2C treatment-induced dedifferentiation of human embryonic stem cell (hESC)-derived cardiomyocytes (CMs) toward ISL1-expressing cells.
Cells induced from hESC-derived CMs by treatment with dimethyl sulfoxide (DMSO) for 60 hr. (A) Phase contrast images showing cell morphology. (B) Immunofluorescence staining of the ISL1 (ISL1, green), and the CM marker cardiac troponin T (TNNT2, red) in the cells. Nuclei were stained by DAPI (4′,6-diamidino-2-phenylindole) and presented in DNA blue. (C) Cytosolic and nuclear areas of the cells. Data are shown as mean ± SD (n = 3 independent experiments, represented as dots). Two-way ANOVA with Dunnett’s multiple comparisons test. ****p < 0.0001. (D) Changes in cell number after 2C treatment. Total cell number, TNNT2+ cell number, and ISL1+ cell number were normalized to the negative control DMSO (NC). Data are shown as mean ± SD (n = 3 independent experiments, represented as dots). Two-way ANOVA with Šidák’s multiple comparisons test. ns, not significant (p > 0.05), ****p < 0.0001. (E) Fraction of ISL1+ cells in TNNT2+ cells. Data are shown as mean ± SD (n = 3 independent experiments, represented as dots). Two-way ANOVA with Šidák’s multiple comparisons test. **p < 0.01. (F) Western blot and quantitative analysis of ISL1 expression in DMSO (NC) or 2C-treated CMs for 60 hr. Data are shown as mean ± SD. Unpaired t test. *p < 0.05. (G) Relative gene expression of embryonic cardiogenesis marker genes (MESP1, ISL1, NR2F2, FUT4, and LEF1), pan-cardiac genes (GATA4, TBX5, and NXK2-5), and CM marker genes (MEF2C, TNNT2, MYL2, and MYL7) in the cells treated by DMSO (NC) or 2C for 60 hours (60 h). Data are shown as mean ± SD (n = 3 independent experiments, represented as dots). Multiple unpaired t tests. ns, not significant (p > 0.05), *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
-
Figure 1—source data 1
PDF file containing original western blots for Figure 1F, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/93405/elife-93405-fig1-data1-v1.zip
-
Figure 1—source data 2
Original files for western blot analysis displayed in Figure 1F.
- https://cdn.elifesciences.org/articles/93405/elife-93405-fig1-data2-v1.zip
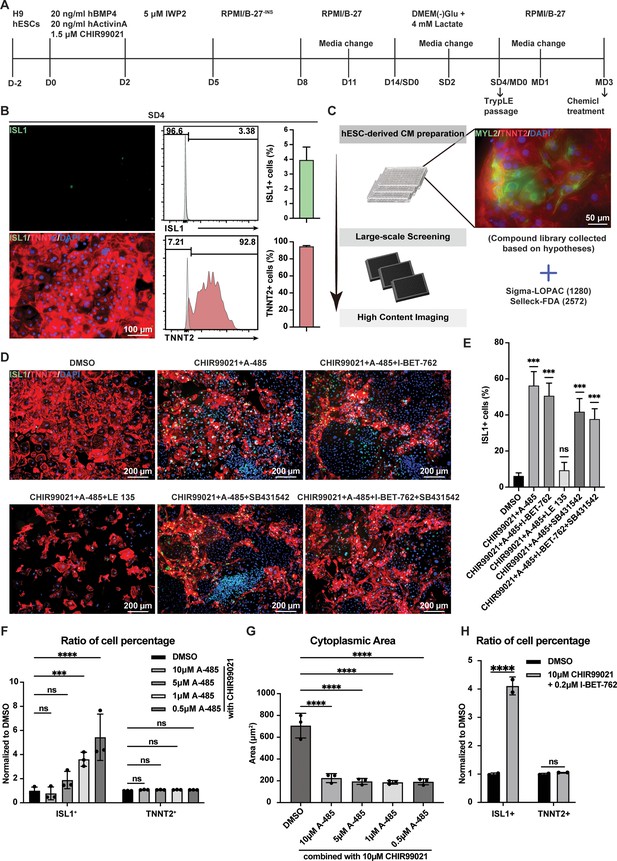
2C-induced reprogramming of ISL1+ cells from human embryonic stem cell (hESC)-derived cardiomyocytes (CMs).
(A) Schematic illustration of optimized protocol to chemically induce and purify CMs differentiated from hESCs. Cells undergoing differentiation from day 0 (D0) to day 8 (D8) were cultured in RPMI1640 basal medium containing B27 without Insulin (RPMI/B-27-INS). After 4 days of selection (SD4) in purification medium containing lactate starting from D14, CMs were maintained in RPMI1640 basal medium containing B27 (RPMI/B-27) from maintenance day 0 (MD0) to maintenance day 4 (MD4). Then CMs were used to induce ISL1-expressing cells by treatment with chemicals. (B) Expression of ISL1 (green) and CM marker TNNT2 (red) was detected by immunofluorescence staining (left) at SD4, and ISL1+ cells and TNNT2+ cells were analyzed by flow cytometry analysis (middle) and presented as their percentage (right). Error bars indicate SD. DAPI (4′,6-diamidino-2-phenylindole) staining labeled nuclei as blue. (C) Scheme of a large-scale chemical content screening by ISL1 immunofluorescence staining using purified hESC-derived CMs. (D) Expression of ISL1 (green) and TNNT2 (red) was shown by immunostaining of CMs treated with various compound combinations for 72 hr. (E) Percentage of ISL1+ cells after 72 hr treatment of CMs with various compound combinations. Data are shown as mean ± SD. Each treatment condition with three technical replicates of values as shown. ns, not significant (p > 0.05), ***p < 0.001, one-way ANOVA with Dunnett’s multiple comparisons test. (F) Titration of the A-485 compound on the effects of CM reprogramming in combination of 10 µM CHIR99021. The ISL1+ and TNNT2+ cell percentages were normalized with the negative control DMSO, respectively. Data are shown as mean ± SD (n = 3 independent experiments, represented as dots). Two-way ANOVA with Dunnett’s multiple comparisons test. ns, not significant (p > 0.05), ***p < 0.001, ****p < 0.0001. (G) Changes in cytoplasmic area of CMs following titrated concentrations of A-485 compounds combined with 10 µM CHIR99021. Data are shown as mean ± SD (n = 3 independent experiments, represented as dots). One-way ANOVA with Dunnett’s multiple comparisons test. ****p < 0.0001. (H) Effects of CM reprogramming induced by the combination of CHIR99021 and I-BET-762. The ISL1+ and TNNT2+ cell percentages were normalized with the negative control DMSO, respectively. Data are shown as mean ± SD (n = 2 independent experiments, represented as dots). Two-way ANOVA with Uncorrected Fisher’s LSD. ns, not significant (p > 0.05), ****p < 0.0001.
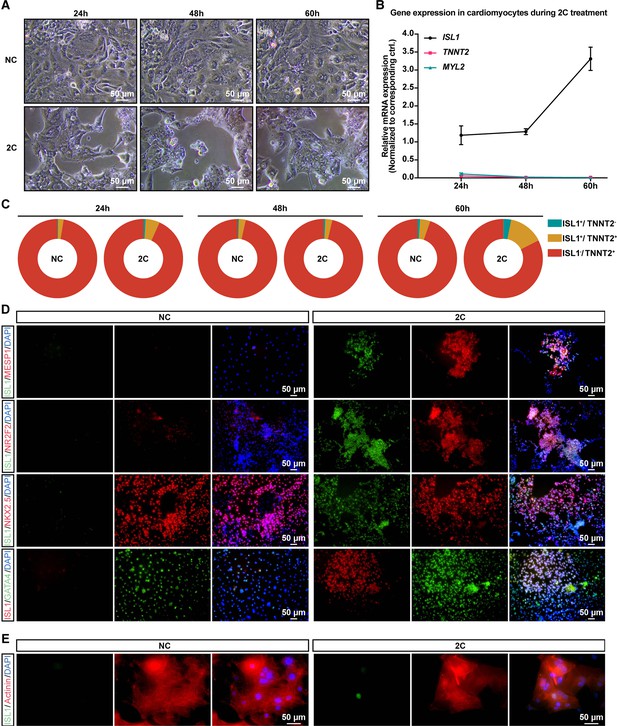
2C treatment gradually induced dedifferentiation of cardiomyocytes (CMs).
(A) Phase contrast images of human embryonic stem cell (hESC)-derived CMs treated by DMSO (NC) or 2C at indicated time points. (B) Gene expression of ISL1, TNNT2, and MYL2 at indicated times during 2C treatment. Data are shown as mean ± SD. Gene expression changes at each time point were normalized with the DMSO at the corresponding time point. (C) Ratios of TNNT2−/ISL1+, TNNT2+/ISL1+, and TNNT2+/ISL1− subpopulations at indicated time points after treatment with 2C or DMSO (NC). (D) Immunostaining for embryonic cardiogenesis marker genes (MESP1 and NR2F2), pan-cardiac genes (NKX2-5 and GATA4), and ISL1 in DMSO (NC) or 2C-treated CMs for 60 hr. DAPI (4′,6-diamidino-2-phenylindole) staining labeled nuclei as blue. ISL1 (green) is used as a marker of dedifferentiation. (E) Immunostaining for ISL1 (green) and CM-specific marker a-Actinin (red) in DMSO (NC) or 2C-treated mature CMs for 60 hr.
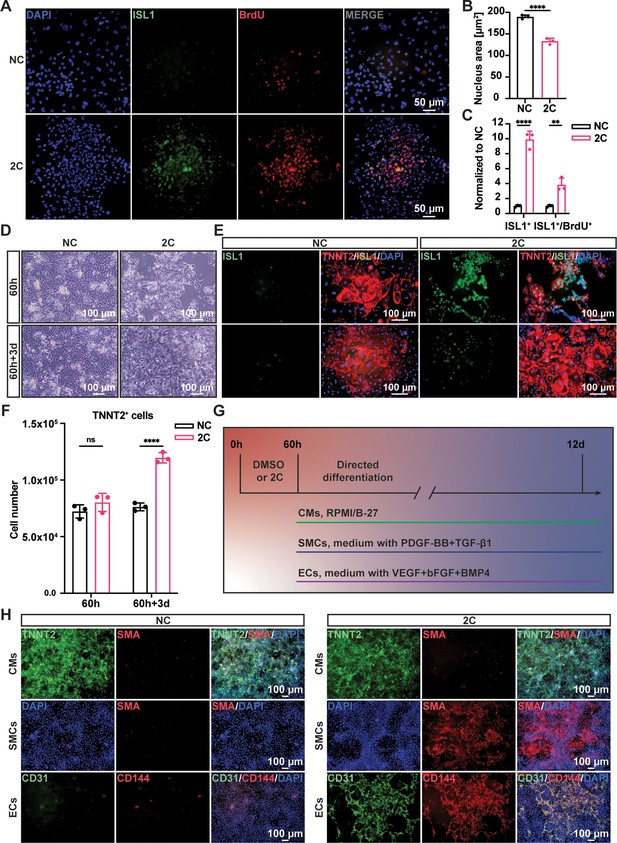
Regenerative ability of 2C-induced regenerative cardiac cells (RCCs).
Immunofluorescence staining (A) and statistical analysis (B, C) of the ISL1 (green) and BrdU (red) double positive RCCs induced from cardiomyocytes (CMs) by treatment with DMSO (NC) or 2C for 60 hr. The ISL1+ cell number and ISL1+/BrdU+ cell number were normalized to the negative control DMSO (NC). DAPI (4′,6-diamidino-2-phenylindole) staining labeled nuclei as blue. Data are shown as mean ± SD (n = 3 independent experiments, represented as dots). Multiple unpaired t tests in (B), ****p < 0.0001. Two-way ANOVA with Šidák’s multiple comparisons test in (C), **p < 0.01, ****p < 0.0001. (D) Phase contrast images of human embryonic stem cell (hESC)-derived CMs treated by DMSO (NC) or 2C for 60 hour (60 h) and subsequently cultured in the absence of 2C for another 3 days (60h+3d). (E) Immunostaining showed the expression of ISL1 (green) and TNNT2 (red) in the cells under the same condition in (D). (F) Statistical analysis of TNNT2+ cell numbers under the same condition in (D). Data are shown as mean ± SD (n = 3 independent experiments, represented as dots). Two-way ANOVA with Šidák’s multiple comparisons test. ns, not significant (p > 0.05), ****p < 0.0001. (G) Schematic diagram of directed differentiation of 2C-induced RCCs toward cardiomyocytes (CMs), smooth muscle cells (SMCs), and endothelial cells (ECs). (H) Immunostaining showed the expression of EC markers (CD31, green and CD144, red), SMC marker (SMA, red), and CM marker (TNNT2, green). DAPI (4′,6-diamidino-2-phenylindole) staining labeled nuclei as blue.
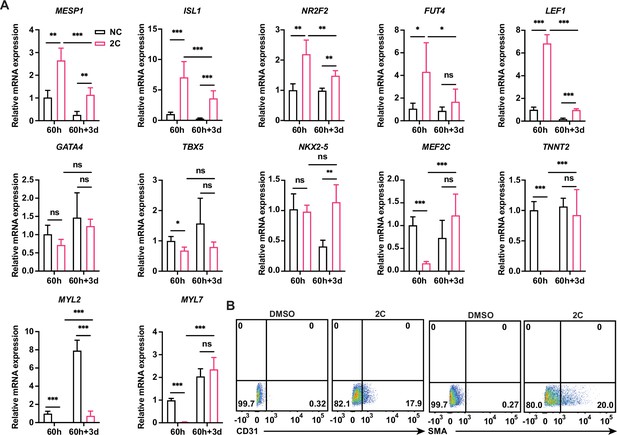
2C-induced regenerative cardiac cells (RCCs) represent the differentiation potential ability.
(A) Relative gene expression of embryonic cardiogenesis marker genes (MESP1, ISL1, NR2F2, FUT4, and LEF1), pan-cardiac genes (GATA4, TBX5, and NXK2-5), and cardiomyocyte (CM) marker genes (MEF2C, TNNT2, MYL2, and MYL7) in the cells treated by DMSO (NC) or 2C for 60 hours (60 h) and subsequently cultured in the absence of 2C for another 3 days (60h+3d). Data are shown as mean ± SD. Multiple unpaired tests. ns, not significant (p > 0.05), *p < 0.05, **p < 0.01, ***p < 0.001. (B) Flow cytometry analysis of the percentage of CD31+ ECs and SMA+ smooth muscle cells (SMCs) in the cells under corresponding differentiation conditions.
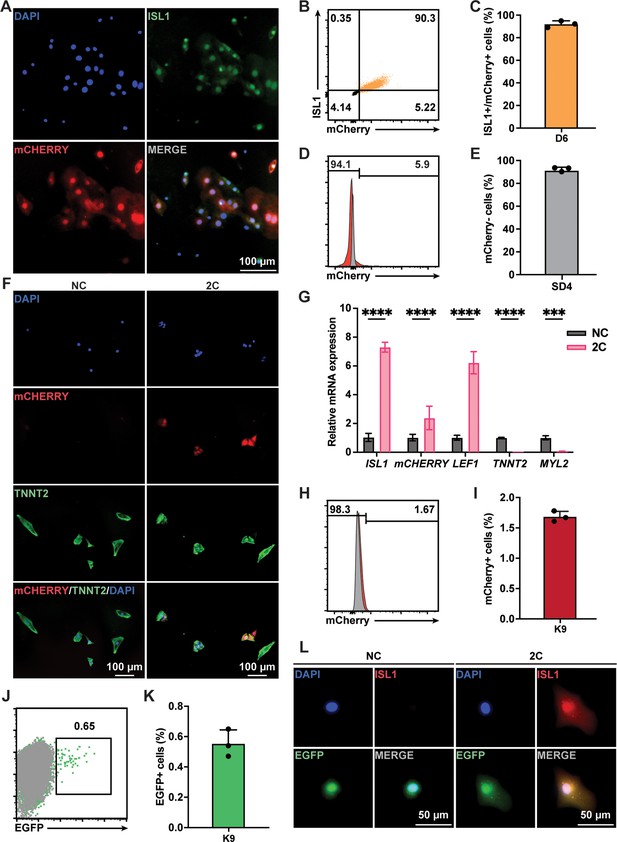
Lineage tracing demonstrated 2C-induced dedifferentiation of TNNT2+ cardiomyocytes (CMs) to ISL1-expressing regenerative cardiac cells (RCCs).
(A) Immunofluorescence images showing expression of endogenous ISL1 (green) and ISL1-mCherry (red) reporter in the cells differentiated from K9 human embryonic stem cell (hESC) KI reporter line at day 6 (D6). DAPI (4′,6-diamidino-2-phenylindole) staining labeled nuclei as blue. (B, C) Flow cytometry analysis of the percentage of mCherry+/ISL1+ cells in the cells differentiated from K9 at D6. (D, E) Flow cytometry analysis of the percentage of mCherry-negative cells at selection day 4 (SD4) in lactate purification medium. (F) Cells induced from mCherry-negative CMs by treatment with or without 2C for 60 hr. Images showing the expression of mCHERRY (red) and TNNT2 (green) in the cells. (G) Relative gene expression of ISL1, mCHERRY, LEF1, TNNT2, and MYL2 in K9-derived mCherry-negative CMs treated with DMSO (NC) or 2C for 60 hr. Data are shown as mean ± SD (n = 2 independent experiments with 4 replicates each). Two-way ANOVA with Šidák’s multiple comparisons test. ***p < 0.001, ****p < 0.0001. (H, I) Flow cytometry analysis of the percentage of mCherry-positive cells induced from mCherry-negative CMs by treatment with or without 2C for 60 hr. Data are shown as mean ± SD (n = 3 independent experiments, represented as dots). Flow cytometric plots showing EGFP-labeled CMs by lineage-tracing of K9-derived mCherry-negative CMs (J), and bar graph showing the percentage of mCherry-negative CMs expressing EGFP (K). Data are shown as mean ± SD (n = 3 independent experiments). (L) Images showing the expression of ISL1 (red) and EGFP (green) in the cells induced from EGFP-positive/mCherry-negative CMs by treatment with or without 2C for 60 hr. DAPI (4′,6-diamidino-2-phenylindole) staining labeled nuclei as blue.
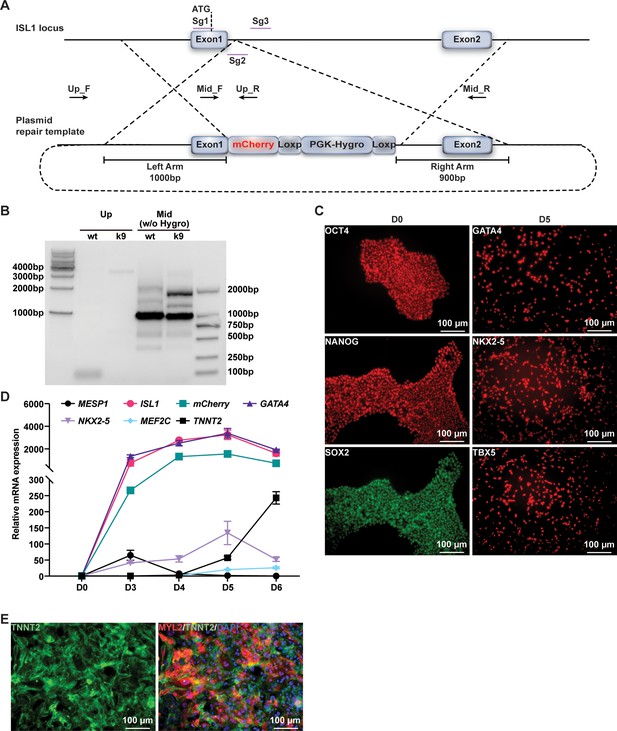
Construction of ISL1mCherry/+knock in H9 human ESC line by CRISPR–Cas9.
(A) A diagram showing the strategy to establish H9 human embryonic stem cell (hESC) line with ISL1-mCherry knock-in reporter. The three single-guide RNAs (Sg1, Sg2, and Sg3) are designed in Exon1 and Intron1 of ISL1, respectively. (B) Genomic PCR confirmed establishment of heterozygous cell lines with ISL1-mCherry knocked in the targeted ISL1 locus. (C) Immunostaining confirmed the expression of ESC marker genes (OCT4, NANOG, and SOX2) and cardiac lineage marker genes (GATA4, NKX2-5, and TBX5) in the cells differentiated from K9 at D0 and D5. (D) Relative expression of genes in the cells differentiated from K9 at indicated time points. Error bars indicate SD. (E) Immunostaining of CM marker genes (TNNT2, green and MYL2, red) in FACS-sorted K9-derived mCherry-negative CMs. DAPI (4′,6-diamidino-2-phenylindole) staining labeled nuclei as blue.
-
Figure 3—figure supplement 1—source data 1
PDF file containing original gel image for Figure 3—figure supplement 1B, indicating the relevant bands.
- https://cdn.elifesciences.org/articles/93405/elife-93405-fig3-figsupp1-data1-v1.zip
-
Figure 3—figure supplement 1—source data 2
Original files for gel image displayed in Figure 3—figure supplement 1B.
- https://cdn.elifesciences.org/articles/93405/elife-93405-fig3-figsupp1-data2-v1.zip
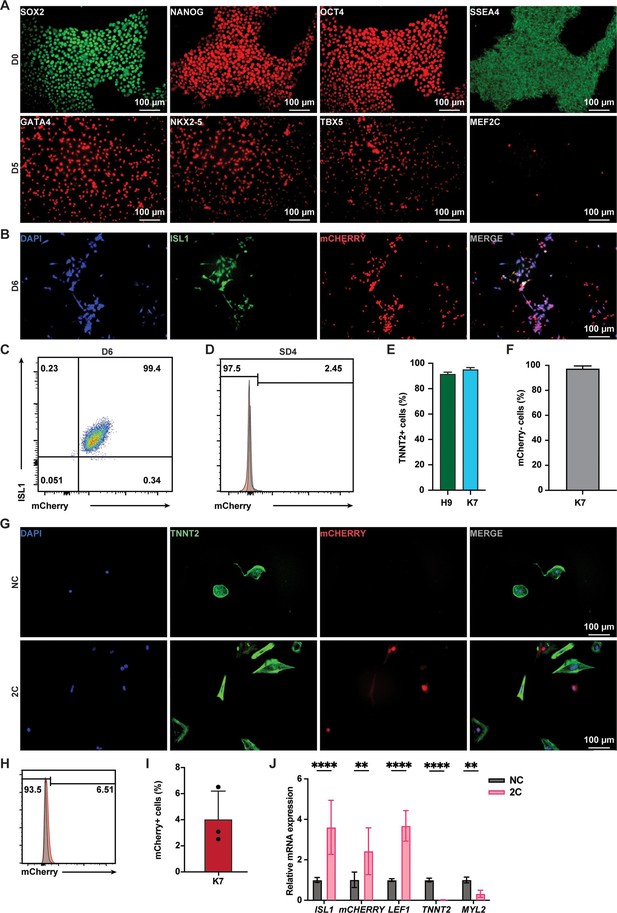
2C-induced K7-derived mCherry-negative cardiomyocytes (CMs) into ISL1/mCherry-positive cells.
(A) Immunostaining of ESC marker genes (SOX2, NANOG, OCT4, and SSEA4) and cardiac lineage marker genes (GATA4, NKX2-5, TBX5, and MEF2C) at D0 and D5 of differentiation of ISL1mCherry/+ HUES7 knock-in (K7) ESC line. (B) Immunostaining showed the co-expression of ISL1 (green) and mCherry (red) in K7-differentiated cells at D6. DAPI (4′,6-diamidino-2-phenylindole) staining labeled nuclei as blue. (C) Fluorescence-activated cell sorting (FACS) analysis showing the percentage of ISL1/mCherry double positive cells in the cells differentiated from K7 at D6. (D) FACS analysis showing the percentage of mCherry+ cells in K7-drived cells cultured in lactate purification media at SD4. (E) Flow cytometric analysis showing the percentages of H9 and K7-derived TNNT2+ CMs at SD4. (F) FACS analysis showing the percentage of K7-derived mCherry-negative cells at SD4. Error bars indicate SD. (G) Immunostaining showed the expression of mCherry and TNNT2 in K7-derived mCherry-negative CMs upon DMSO (NC) or 2C treatment for 60 hr. DAPI staining labeled nuclei as blue. (H, I) Flow cytometry analysis of the percentage of mCherry-positive cells dedifferentiated from K7 human embryonic stem cell (hESC) KI reporter line derived mCherry-negative CMs. Data are shown as mean ± SD (n = 3 independent experiments, represented as dots). (J) Relative gene expression of ISL1, mCHERRY, LEF1, TNNT2, and MYL2 in K7-derived mCherry-negative CMs treated with DMSO (NC) or 2C for 60 hr, respectively. Data are shown as mean ± SD (n = 2 independent experiments with 4 replicates each). Two-way ANOVA with Šidák’s multiple comparisons test. **p < 0.01, ****p < 0.0001.
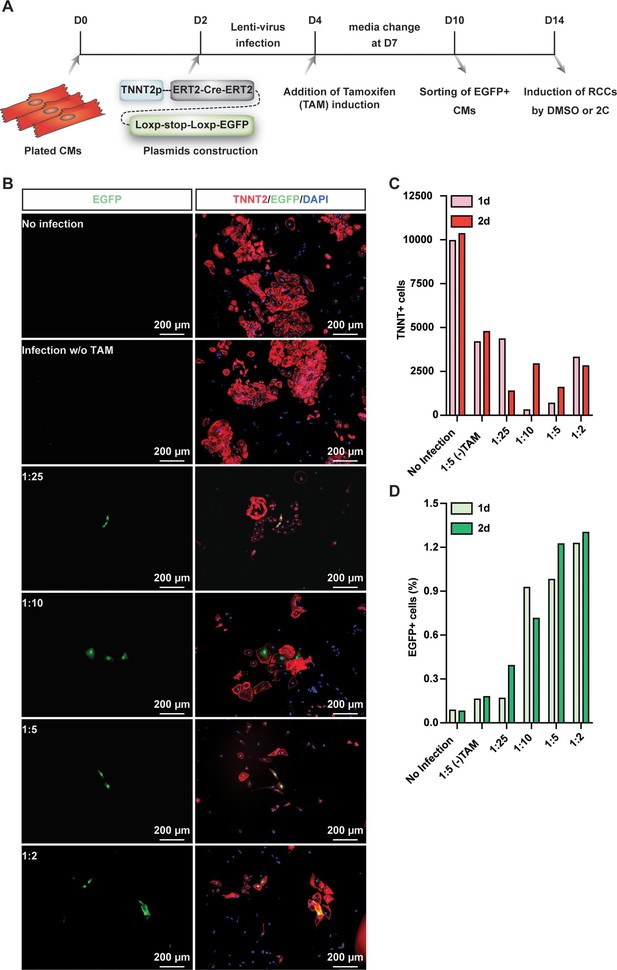
2C-induced ISL1 expression in cardiomyocytes (CMs) with EGFP labeled by lineage tracing.
(A) Scheme of lineage tracing to label EGFP in CMs. (B) Immunostaining showed the expression of EGFP in TNNT2+ CMs infected with or without different titers of virus. DAPI (4′,6-diamidino-2-phenylindole) staining labeled nuclei as blue. (C) The number of TNNT2+ cells alive after infection with or without different titers of virus. (D) The percentage of EGFP+ cells in TNNT2+ CMs infected with or without different titers of virus.
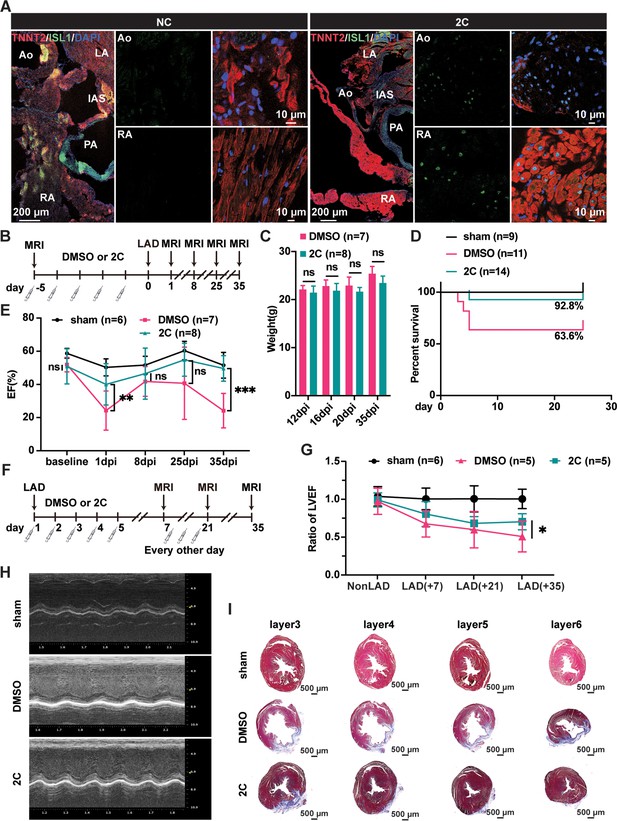
Heart regeneration via 2C-induced dedifferentiation of cardiomyocytes (CMs).
(A) Immunofluorescence staining of ISL1 (green) and TNNT2 (red) in cross-sectioned hearts from 2C or vehicle (DMSO)-treated (NC) adult 129SvJ mice. Ao, aorta. PA, pulmonary artery. LA, left atrial. RA, right atrial. IAS, interatrial septum. DAPI (4′,6-diamidino-2-phenylindole) staining labeled nuclei as blue. (B) Schematic illustration of the method used to examine the prophylactic effect of 2C in 129SvJ mice post myocardial infarction (MI). (C) Body weight of mice pre-treated with vehicle (DMSO) or 2C as shown in (B) at day 12, day 16, day 20, and day 35 after MI (dpi). Error bars represent SD. ns, not significant (p > 0.05). (D) Survival curve of sham-operated mice and mice pre-treated with vehicle (DMSO) or 2C as shown in (B), at indicated time points before or after MI. (E) Ejection fraction (EF) of sham-operated mice and mice pre-treated with vehicle (DMSO) or 2C as shown in (B), before MI (baseline) or at day 1, day 8, day 25, and day 35 after MI (dpi). Data are shown as mean ± SD. Two-way ANOVA with Tukey’s multiple comparisons test. ns, not significant (p > 0.05), **p < 0.01, ***p < 0.001. (F) Schematic illustration of the method used to examine therapeutic effect of 2C in the 129SvJ mice post MI. (G) Serial fMRI measurements showing the cardiac function from sham-operated mice and mice treated with vehicle (DMSO) or 2C at as shown in (F). Data are shown as mean ± SD. Two-way ANOVA with Tukey’s multiple comparisons test. *p < 0.05. (H) Echocardiography of sham-operated mice and mice treated with vehicle (DMSO) or 2C as shown in (F) at day 35 post MI. (I) Masson staining of serial transverse sections of hearts from sham-operated mice and mice treated with vehicle (DMSO) or 2C as shown in (F) at day 35 post MI.
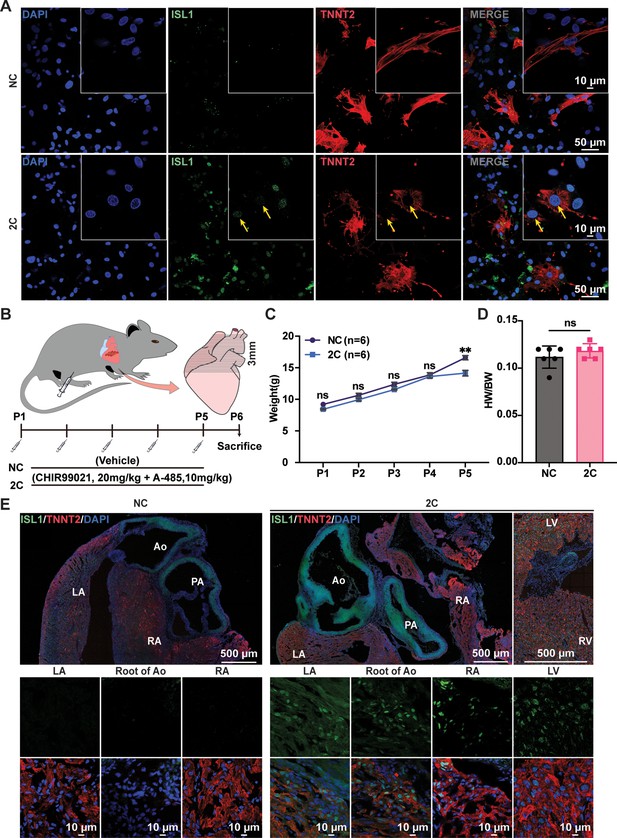
2C-induced expression of ISL1 in neonatal rats cardiomyocytes (CMs) in vitro and in vivo.
(A) Immunostaining of Isl1 (green) and Tnnt2 (red) in primary neonatal rats ventricular CMs (NRVCMs) treated with DMSO (NC) or 2C for 60 hr. In 2C-treated cells, cells co-expressing Isl1 and Tnnt2 were indicated by yellow arrows. (B) Schematic diagram of administration of neonatal SD rats with vehicle (DMSO, NC) or 2C (20 mg/kg CHIR99021 and 10 mg/kg A-485) from the day of birth (P1). Body weights (C) in neonatal rats by administration with DMSO (NC) or 2C at indicated time points and the ratio (D) of heart to body weights (HW/BW) under the same condition at day 6. Data are shown as mean ± SD (n = 6). Two-way ANOVA with Šidák’s multiple comparisons test in (C) and unpaired t test in (D). ns, not significant (p > 0.05), **p < 0.01. (E) Immunofluorescence staining of Isl1 (green) and Tnnt2 (red) in cross-sectioned hearts from neonatal rats with the same treatment in (B). DAPI (4′,6-diamidino-2-phenylindole) staining labeled nuclei as blue. Ao, aorta; PA, pulmonary artery; LA, left atrial; RA, right atrial; LV, left ventricle; RV, right ventricle.
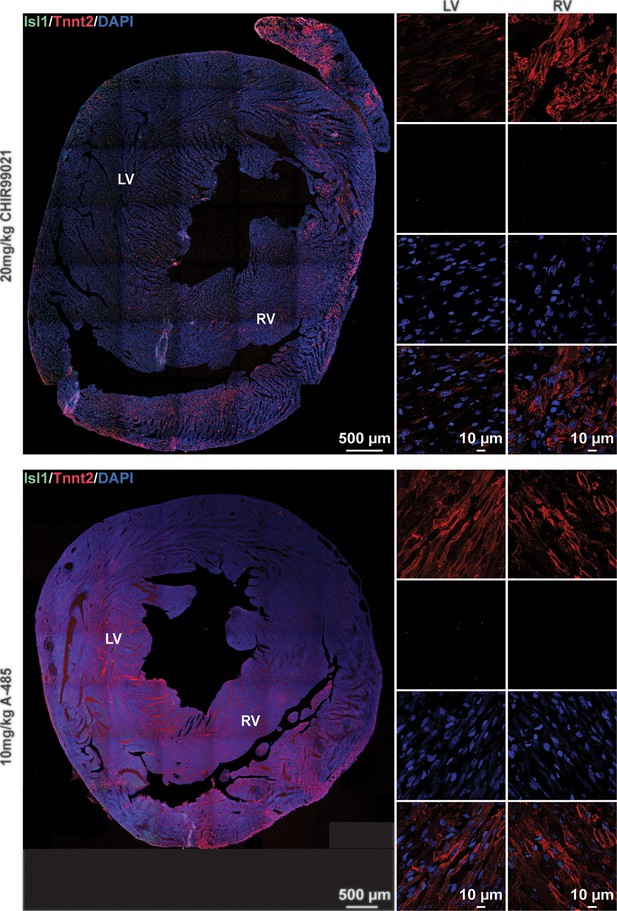
Administration of CHIR99021 or A-485 alone cannot induce ISL1 expression in neonatal rat cardiomyocytes (CMs) in vivo.
Immunofluorescence staining of Isl1 (green) and Tnnt2 (red) in cross-sectioned hearts from neonatal rats with the same treatment in (Figure 4—figure supplement 1B). DAPI (4′,6-diamidino-2-phenylindole) staining labeled nuclei as blue. LV, left ventricle; RV, right ventricle.
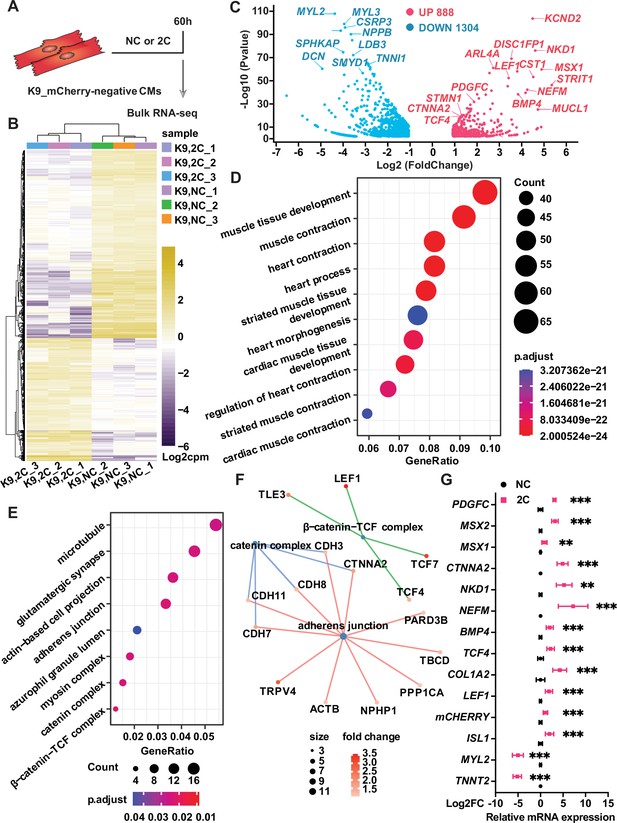
Bulk RNA-seq of analysis of 2C-treated ISL1/mCherry-negative cardiomyocytes (CMs).
(A) Scheme of bulk RNA-seq analysis of K9-derived mCherry-negative CMs with DMSO (NC) or 2C treatment for 60 hr. (B) Heatmap of differentially expressed genes (DEGs) in ISL1/mCherry-negative CMs treated with DMSO (NC) or 2C for 60 hr. (C) Volcano plot showing genes significantly changed by 60 hr of 2C treatment. Gene ontology (GO) analysis of downregulated (D) and upregulated (E) genes in ISL1/mCherry-negative CMs by 2C treatment for 60 hr, compared to DMSO (NC) treated cells. (F) Plotting GO terms of upregulated genes by 2C treatment with cnetlpot. (G) Relative expression fold-changes of indicated genes in K9-derived ISL1/mCherry-negative CMs by 60 hr of DMSO (NC) or 2C treatment. Data are shown as mean ± SD. Multiple unpaired t tests. **p < 0.01, ***p < 0.001.
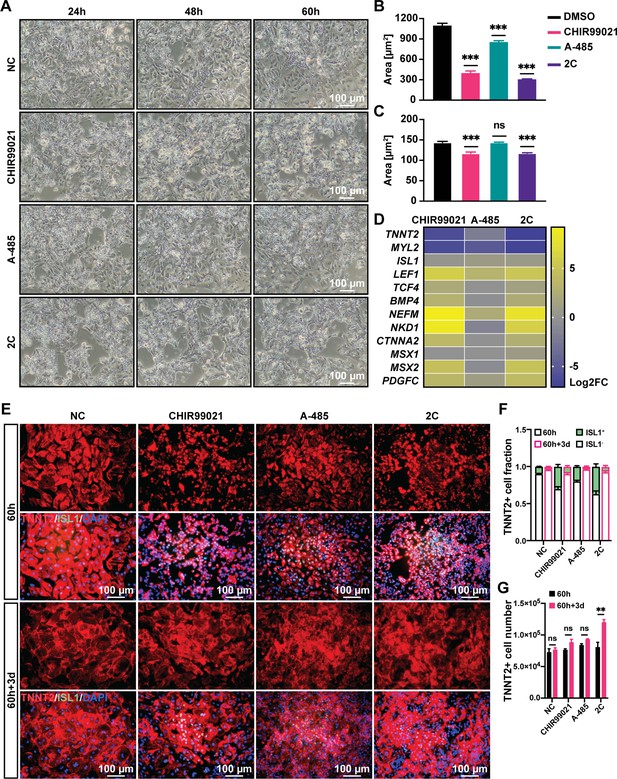
The individual and cooperative effects from CHIR99021 and A-485 on the induction of H9 human embryonic stem cell (hESC)-derived cardiomyocytes (CMs) into regenerative cardiac cells (RCCs).
(A) Phase contrast images of hESC-derived CMs treated by DMSO (NC), CHIR99021, A-485, or 2C at indicated time points. Cytosolic (B) and nuclear (C) areas of the cells treated by DMSO (NC), CHIR99021, A-485, or 2C at indicated time points. Data are shown as mean ± SD (n = 2 independent experiments with 3 replicates each). One-way ANOVA with Dunnett’s multiple comparisons test. ns, not significant (p > 0.05), ***p < 0.001. (D) Heatmap illustration showing the fold-changes of indicted marker genes’ expression by small molecule treatment, which were measured using quantitative real-time PCR (qRT-PCR). (E) Immunostaining showed the expression of ISL1 (green) and TNNT2 (red) in the cells treated by DMSO (NC), CHIR99021, A-485, or 2C for 60 hours (60 h) and subsequently cultured in the absence of 2C for another 3 days (60h+3d). DAPI (4′,6-diamidino-2-phenylindole) staining labeled nuclei as blue. (F) The ratio of ISL1+ cells in TNNT2+ cells at indicated time points in (E). Data are shown as mean ± SD (n = 2 independent experiments with 3 replicates each). Two-way ANOVA with Šidák’s multiple comparisons test. (G) The number of TNNT2+ cells in the cells treated with the same condition in (E) at indicated time points. Data are shown as mean ± SD (n = 2 independent experiments with 3 replicates each). Multiple unpaired t tests. ns, not significant (p > 0.05), **p < 0.01.
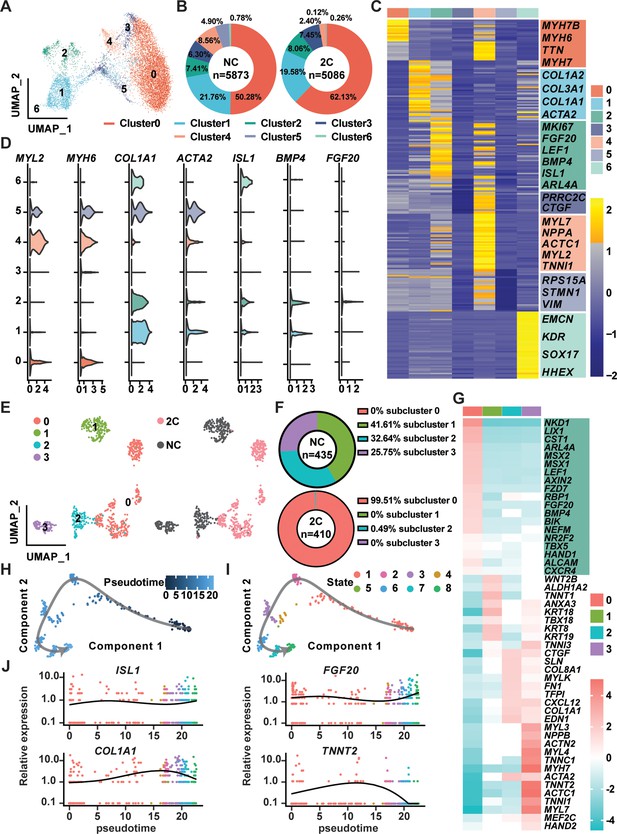
Single-cell RNA-seq of 2C-treated mCherry-negative cardiomyocytes (CMs).
(A) UMAP analysis showing seven clusters in cells induced from K9-derived mCherry-negative CMs by treatment with DMSO (NC) and 2C for 60 hr. (B) The percentage of cells in the seven indicated clusters, following DMSO (NC) or 2C treatment. (C) Heatmap showing the differentially expressed genes in the cells from seven indicated clusters. The representative marker genes of seven indicated clusters were listed on the right. (D) Violin plots showing the expression levels of marker genes of CMs (MYL2, MYH6), intermediate cells (ICs) (COL1A1, ACTA2), and regenerative cardiac cells (RCCs) (ISL1, BMP4, FGF20) among cells from seven indicated clusters. (E) UMAP analysis showing the second-level clustering of cluster 2 into four subclusters (left), which exhibited dramatic distinction under condition of 2C or NC (right). (F) The percentage of cells in the four indicated subclusters within cluster 2, following DMSO (NC) or 2C treatment. (G) Heatmap showing the differentially expressed genes among cells from four subclusters of cluster 2. Genes related to RCCs are highlighted in the green blocks on the right. Pseudotime trajectory showing changes across various cell states upon 2C treatment, which were presented with different developmental pseudotime points (H) and cell states (I). (J) Curves showing the dynamic expression of representative genes of RCCs (ISL1, FGF20), ICs (COL1A1), and CMs (TNNT2) along indicated pseudotime points.
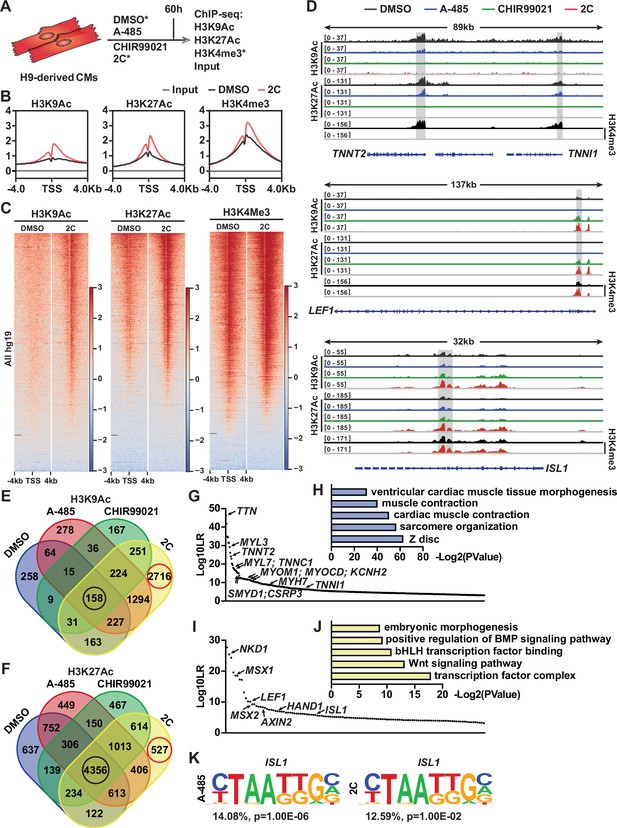
Chromatin immunoprecipitation-sequencing (ChIP-seq) analyses of chemical-treated H9 human embryonic stem cell (hESC)-derived cardiomyocytes (CMs).
(A) Schematic illustration of ChIP-seq analysis of H9-derived CMs subjected to DMSO, A-485, CHIR99021, or 2C treatment for 60 hr. (B) Average ChIP-seq signal profiles showing the indicated histone modifications around the transcription start site (TSS) in the input and ChIP samples prepared from DMSO and 2C-treated cells. (C) Heatmap showing the whole-genome wide distribution of H3K9Ac, H3K27Ac, and H3K4me3 peaks within a range of ±4 kb from TSSs in the cells treated with DMSO or 2C for 60 hr. (D) H3K9Ac and H3K27Ac peaks surrounding CM genes (TNNT2 and TNNI1) and regenerative cardiac cell (RCC) genes (LEF1 and ISL1) in the cells treated with DMSO, A-485, CHIR99021, or 2C for 60 hr, and H3K4me3 peaks surrounding the same genes in the cells treated by DMSO or 2C. The y-axis represents the number of counts. (E, F) Venn diagram showing the number of annotated genes with H3K9Ac or H3K27Ac enrichment in the cells treated with DMSO, A-485, CHIR99021, or 2C for 60 hr. Red circles indicate the number of genes with unique H3K9Ac or H3K27Ac enrichment induced by 2C treatment; black circles indicate the number of genes with H3K9Ac or H3K27Ac enrichment unaffected by any chemical treatment. The annotated genes with the most significant changes in H3K9Ac enrichment following treatment with DMSO (G) or 2C (I) were ranked by Log10LR and analyzed by gene ontologies (GOs) (H, J), respectively. (K) ISL1-binding motifs identified from the cells treated with A-485 or 2C.
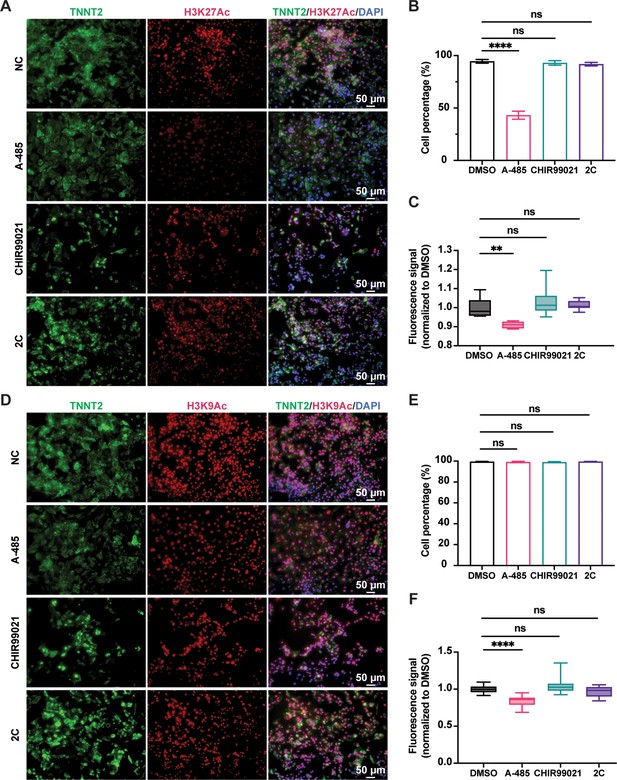
H3K27Ac and H3K9Ac levels regulated by CHIR99021 and A-485 individually or in combination.
Cells induced from H9 human embryonic stem cell (hESC)-derived cardiomyocytes (CMs) by treatment with DMSO (NC), CHIR99021, A-485, or 2C for 60 hr. (A–C) H3K27Ac levels in the cells treated with indicated small molecules. Immunostaining (A) and quantitative analysis of H3K27Ac levels in the TNNT2+ CMs (B, C). H3K9Ac levels in the cells treated with indicated small molecules. Immunostaining (D) and quantitative analysis of H3K9Ac levels in the TNNT2+ CMs (E, F). DAPI (4′,6-diamidino-2-phenylindole) staining labeled nuclei as blue. CMs stained by TNNT2 (green). Data are shown as mean ± SD (n = 2 independent experiments with 4 replicates each). One-way ANOVA with Dunnett’s multiple comparisons test. ns, not significant (p > 0.05), **p < 0.01, ****p < 0.0001.
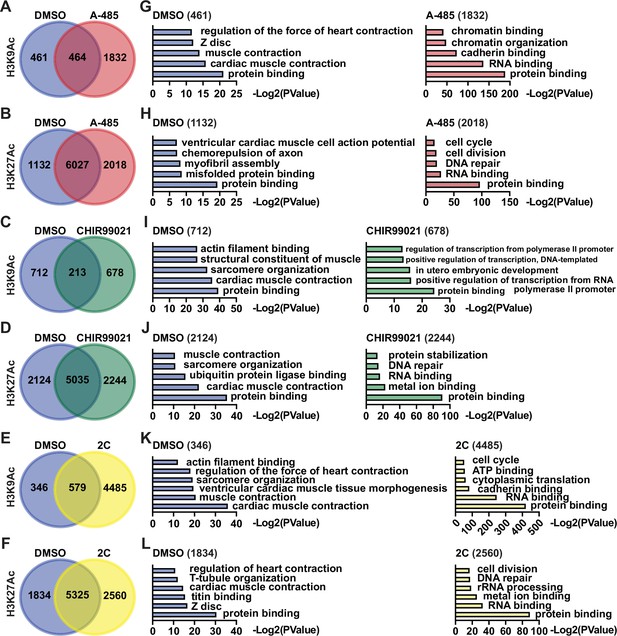
Chemical treatment alters the expression of signature genes by regulating H3K27Ac and H3K9Ac in proximity.
Gene ontology (GO) analyses of genes associated with H3K9Ac and H3K27Ac enrichments in the cells treated with DMSO, A-485, CHIR99021, or 2C for 60 hr. Venn diagram showing the number of genes enriched by H3K9Ac and H3K27Ac after treatment with DMSO, A-485 (A, B), CHIR99021 (C, D), or 2C (E, F). GO analyses of genes with H3K9Ac and H3K27Ac enrichments observed following treatment with DMSO, A-485 (G, H), CHIR99021 (I, J), or 2C (K, L).
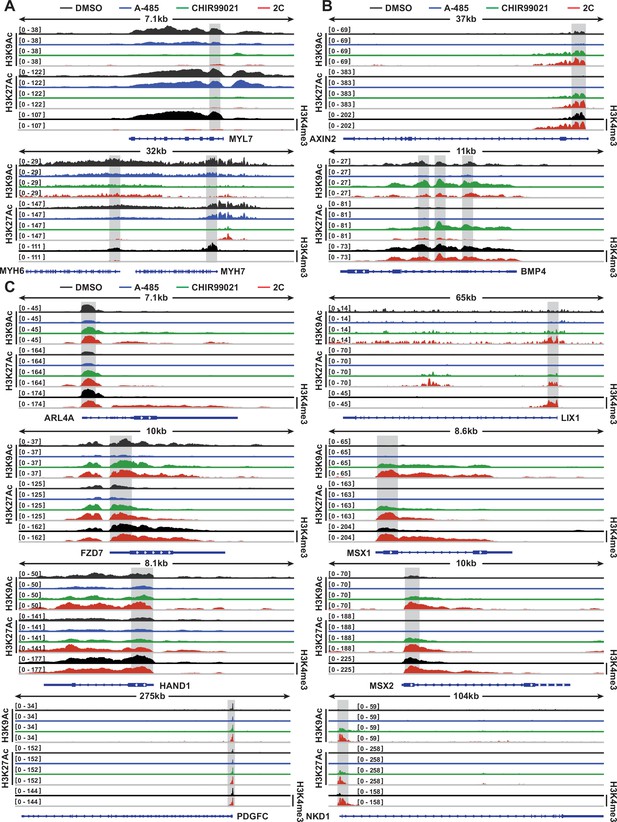
Chemical treatment differentially altered the peaks of H3K9Ac and H3K27Ac on cardiomyocyte (CM) and regenerative cardiac cell (RCC) genes.
(A) Peaks of H3K9Ac and H3K27Ac on CM genes in the cells treated with DMSO, A-485, CHIR99021, or 2C, and H3K4me3 peaks on the same genes in the cells treated with DMSO or 2C. (B, C) Peaks of H3K9Ac and H3K27Ac on RCC genes in the cells treated with DMSO, A-485, CHIR99021, or 2C, and H3K4me3 peaks on the same genes in the cells treated with DMSO or 2C. The y-axis represents the number of counts.
Videos
Contracted cardiomyocytes (CMs) at SD4.
Human embryonic stem cell (hESC)-derived CMs at day 4 (SD4) after lactate selection.
Contracted cardiomyocytes (CMs) at 60h+3d-2C.
Human embryonic stem cell (hESC)-derived CMs treated by 2C for 60 hours (60 h) and subsequently cultured in the absence of 2C for another 3 days (60h+3d).
Additional files
-
Supplementary file 1
Table of compound library collected based on hypotheses.
The compounds from the lab’s proprietary library, along with their respective targets and working concentrations, are provided in the table.
- https://cdn.elifesciences.org/articles/93405/elife-93405-supp1-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/93405/elife-93405-mdarchecklist1-v1.docx





