Polysaccharide breakdown products drive degradation-dispersal cycles of foraging bacteria through changes in metabolism and motility
Figures
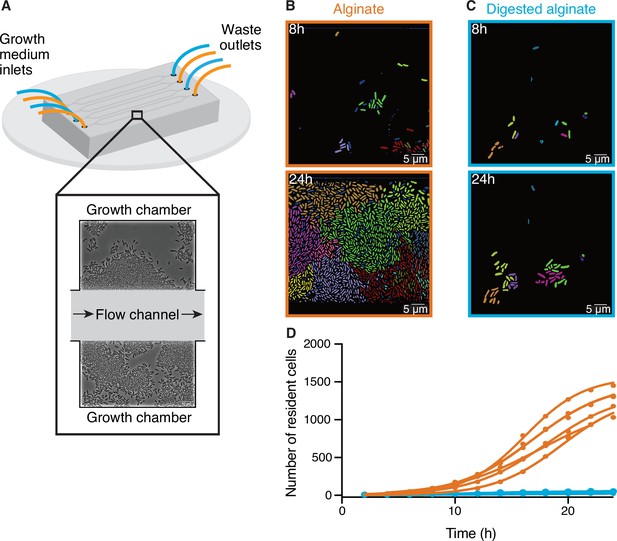
Large cell groups form on alginate but not on digested alginate.
(A) Schematic representation of the setup of the microfluidic experiments. (B and C) Representative images at different time points of V. cyclitrophicus ZF270 cells growing in microfluidic chambers, described in detail by Dal Co et al., 2020, with (B) alginate medium or (C) digested alginate medium, both in their soluble form (not visible). Cells are false-colored according to their lineage identities based on cell segmentation and tracking over 24 hr. Cells without identified progenitors are colored in dark blue. See Figure 1—video 1 (alginate) and Figure 1—video 2 (digested alginate) for time-lapse videos. (D) Cell numbers within microfluidic chambers supplied with alginate (orange) are substantially higher than cell numbers within microfluidic chambers supplied with digested alginate (blue) (Logistic growth regression for alginate: R2=0.99, maximal number of cells = 1217–1564, k=0.24–0.38 hr–1; for digested alginate: R2=0.86–0.97, maximal number of cells = –100, k=0.07–0.4 hr–1). Circles indicate the number of cells present at a given time point in each chamber (nchambers = 7). Data for chambers with alginate originate from D’Souza et al., 2023a. Lines are fits of a logistic growth regression line for each condition.
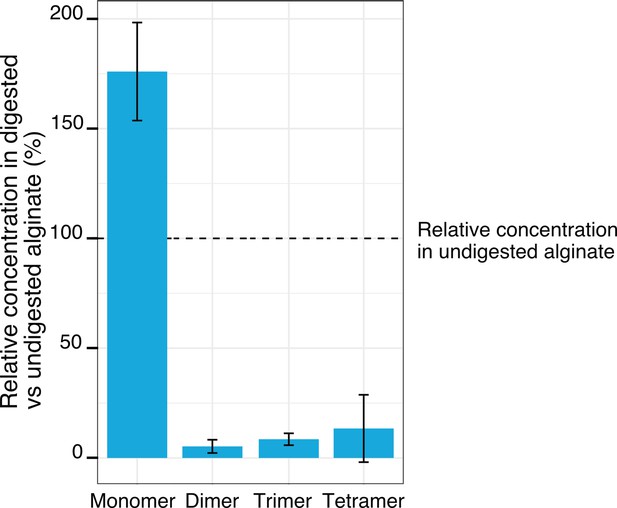
Relative concentrations of the breakdown products of alginate after treatment with commercial alginate lyases.
LC-MS measurements of the digested and undigested alginate media, comparing the abundance of monomers, dimers, trimers, and tetramers in the digested alginate medium to the undigested one. The digestion was achieved by incubation with commercial alginate lyases for 48 hr. Bar heights depict the mean of four biological replicates whereas whiskers depict the standard deviation.
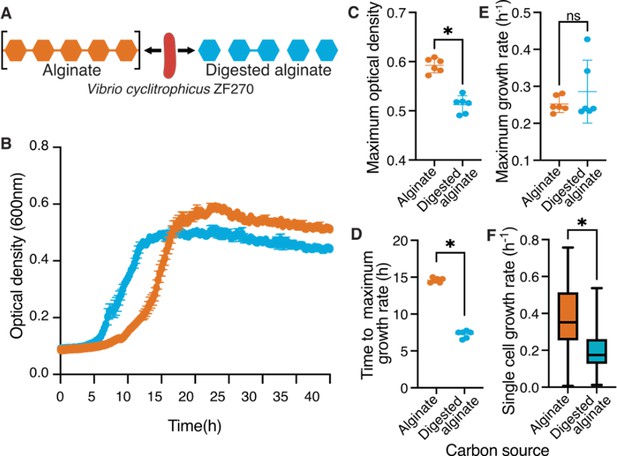
Polymeric alginate increases lag times and yield of Vibrio cyclitrophicus ZF270 populations.
(A) Vibrio cyclitrophicus ZF270 was grown in microwell plates on 0.1% (w/v) polymeric (alginate) or on 0.1% digested alginate as a sole carbon source. (B) Bacterial growth measured for 40 hr using optical density (OD) at 600 nm. (C) Maximum OD600 on polymeric alginate (orange) or digested alginate (blue). Note that while the OD curves do not reach the exact same OD, plating cells on agar plates at 36 hr resulted in the same number of colonies (Figure 1—figure supplement 3), indicating that the OD readings may be affected by the polysaccharide in the media. (D) Time to achieve maximal growth rates (lag time) and (E) maximum growth rates on polymeric alginate (orange) and digested alginate (blue). Circles indicate individual measurements, whereas horizontal lines indicate the mean and whiskers the confidence intervals (CI) of six replicate populations. (F) Distribution of single cell growth rates of cells growing within microfluidic growth chambers on polymeric alginate (orange) or digested alginate (blue). Median growth rates were measured for each growth chamber for every 2 hr interval. Boxes extend from the 25th to 75th percentiles, whiskers indicate the 10th and 90th percentiles of median growth rates, and horizontal lines mark the median growth rates. Asterisks or ns indicate statistically significant or non-significant comparisons, respectively (independent samples t-test, in C: p<0.0001, t=8.674, n=6; in D: p=0.34, t=0.98, n=6; in E: p<0.0001, t=30.73, n=6; in F: Mann-Whitney Test, p<0.0001).
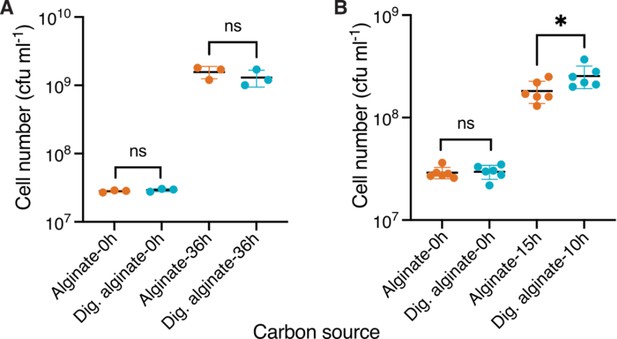
Cell counts of V. cyclotrophicus ZF270 on alginate and digested alginate measured by plating assay.
(A) Cells were grown in shaking flasks and the yield was measured at the start of the experiment (0 hr) and after 36 hr by plating on Marine Agar plates. Shown are colony forming units ml –1 (cfu ml–1) formed 24 hr after plating. Circles indicate individual measurements whereas horizontal lines show the mean and whiskers represent the confidence intervals (CI) of three replicate populations. Cell numbers were statistically non-significant, depicted by ns, amongst groups (independent samples t-test, 0 hr: p=0.33, t=1.093, n=3; 36 hr: p=0.39, t=0.95, n=3). (B) Cells were grown on 0.1% (w/v) polymeric or on 0.1% oligomeric alginate in shaking flasks and the yield was measured at the start of the experiment (0 hr) and when harvesting for RNA extraction, that is during exponential phase [based on Figure 1: 10 hr for digested alginate (OD: 0.34) and 15 hr for alginate (OD: 0.33)] by plating on Marine Agar plates. Shown are colony forming units ml –1 (cfu ml–1) formed 24 hr after plating. Circles indicate individual measurements, whereas horizontal lines show the mean and whiskers represent the confidence intervals (CI) of three replicate populations. Cell numbers were statistically non-significant, depicted by ns, amongst groups (independent samples t-test, 0 hr: p<0.64, R2=0.006, t=0.25, n=6; 36 hr: p=0.04, R2=0.34, t=2.318, n=3). Dig. alginate: digested alginate.
Time-lapse video of Vibrio cyclitrophicus ZF270 cells within a representative microfluidics chamber fed with 0.1% alginate as the sole carbon source.
Images were captured every 8 min. Cells are false colored based on the identity of their progenitor cells. Cells whose divisional history cannot be tracked are shown in blue. The scale bar corresponds to 10 µm.
Time-lapse video of Vibrio cyclitrophicus ZF270 cells within a representative microfluidics chamber fed with 0.1% digested alginate as the sole carbon source.
Images were captured every 8 min. Cells are false colored based on the identity of their progenitor cells. Cells whose divisional history cannot be tracked are shown in blue. The scale bar corresponds to 10 µm.
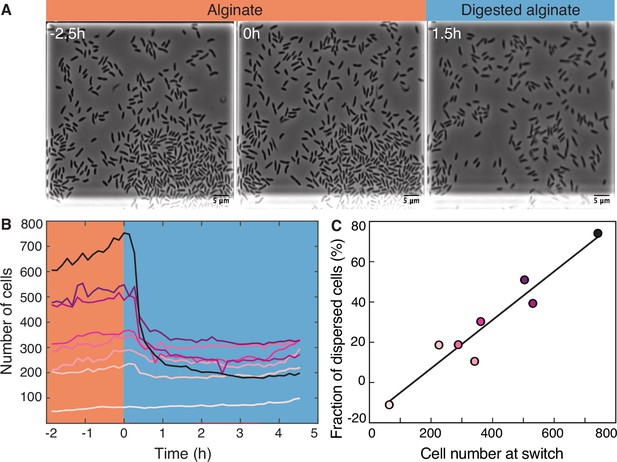
Transition from alginate to digested alginate triggers density-dependent dispersal of cells.
(A) Representative time-lapse images of V. cyclitrophicus ZF270 cells (phase contrast microscopy) in microfluidic growth chambers that were initially exposed to alginate and then switched to digested alginate. (B) Number of cells in different chambers over time, each chamber indicated by a unique color (n=8). The carbon source is indicated by the colored background (orange: alginate; blue: digested alginate). See Figure 2—video 1 for a time-lapse video. (C) Positive relationship between the number of cells in the microfluidic growth chamber at the time of the switch and the fraction of cells that disperse after the switch. Each circle represents one growth chamber with colors corresponding to (B), and the line depicts a linear regression fit (R2=0.92, slope = 0.12).
Time-lapse video of Vibrio cyclitrophicus ZF270 cells within a representative microfluidics chamber fed with 0.1% alginate and then switched to 0.1% digested alginate as sole carbon sources.
Timings of the switch and carbon sources are indicated on the images. Images were captured every 8 min. The scale bar corresponds to 10 µm.
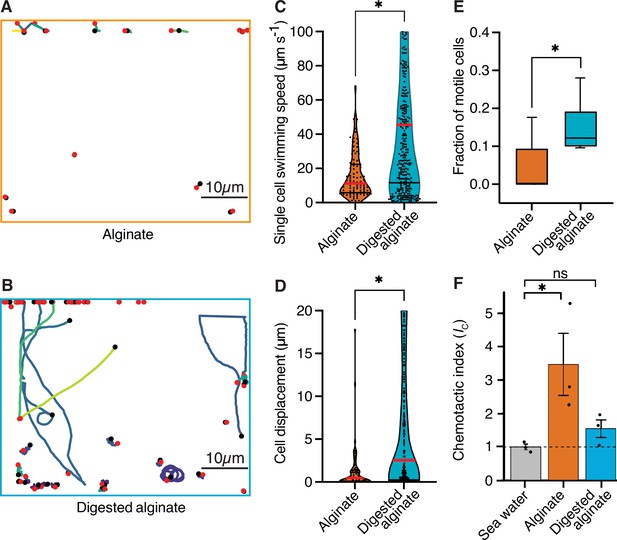
Cells are more motile on digested alginate than alginate and show chemotaxis towards alginate.
(A and B) Spatial trajectories of cells supplied with (A) alginate or (B) digested alginate in representative microfluidic growth chambers are shown. Black points mark the starting point of each trajectory, pink points mark the end point of each trajectory, and colored lines mark the trajectories of cells. (C) Distributions of the mean single-cell swimming speeds (Nested t-test, p-value <0.0007, t=4.803, df = 10, ncells = 86 vs 375 in nchambers = 5) are shown. (D) Distributions of cell displacement over the course of a trajectory (Nested t-test test, p-value <0.0131, t=4.39, df = 10, ncells = 86 vs 375, and nchambers = 5) are shown. In (C) and (D) the red horizontal lines indicate the mean while black lines depict the 25th and 75th quartiles of the distribution. (E) The mean fraction of motile cells in each chamber, where motile cells are defined as cells with displacement greater than 1 µm (Mann-Whitney test on the means of five growth chambers, p-value = 0.034). In C, D, and E, each chamber was considered as an independent replicate. (F) Chemotactic index (IC) quantified by In Situ Chemotaxis Assay (ISCA) (Tukey multiple comparisons of means, 95% family-wise confidence levels as error bars, p-value <0.05, n=3). Asterisks indicate statistically significant differences. See Figure 3—video 1 and Figure 3—video 1 for time-lapse videos of swimming cells.
High frame rate (125 Hz, i.e. frames s–1) time-lapse video of Vibrio cyclitrophicus ZF270 cells within a representative microfluidics chamber fed with 0.1% alginate as the sole carbon source.
The scale bar corresponds to 10 µm.
High frame rate (125 Hz, i.e., frames s–1) time-lapse video of Vibrio cyclitrophicus ZF270 cells within a representative microfluidics chamber fed with 0.1% digested alginate as the sole carbon source.
The scale bar corresponds to 10 µm.
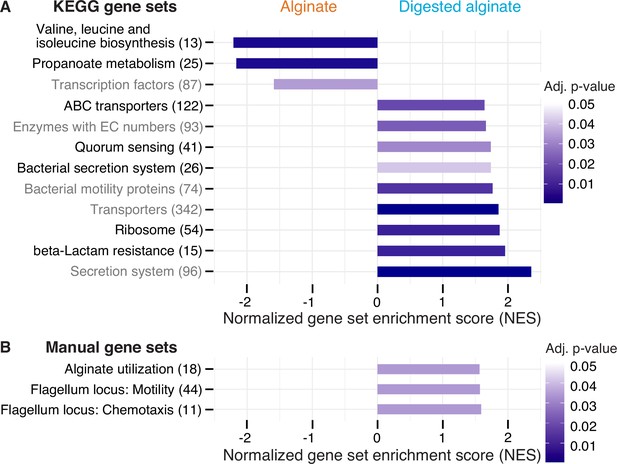
Twelve functional gene sets are enriched in genes with increased or decreased expression in cells grown on digested alginate.
Gene set enrichment analysis (GSEA) with (A) all KEGG pathways and KEGG BRITE categories as gene sets or with (B) a custom alginate utilization, flagellar assembly, and flagellum-driven chemotaxis gene set was performed comparing the gene counts of the transcriptome of V. cyclitrophicus ZF270 cultures grown on digested alginate and alginate (six replicates each). Gene sets with a positive enrichment score were enriched with genes with higher expression in cells grown on digested alginate relative to cells grown on alginate (BH-adjusted p-value <0.05), whereas gene sets with negative enrichment scores were significantly enriched with genes with decreased expression on digested alginate. The number in brackets indicates the number of genes with unique K number per gene set (A) and the number of genes per gene set (B) within the V. cyclitrophicus ZF270 genome.
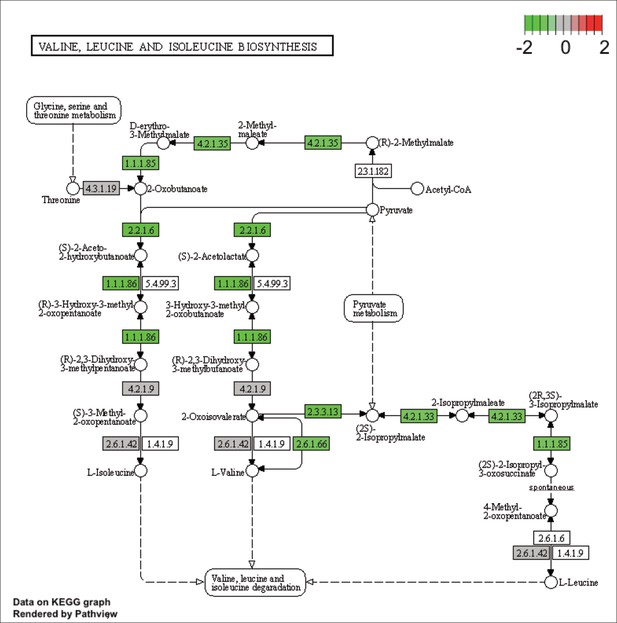
KEGG map of the significantly enriched KEGG pathway for valine, leucine and isoleucine biosynthesis.
The labeled KEGG maps were created with the R package pathview v.1.35.0 for each KEGG pathway that was identified as a significantly enriched gene set by GSEA (see Materials and methods). The color represents the log2 fold change of gene expression in V. cyclitrophicus ZF270 cells grown on digested alginate compared to alginate (green-grey-red color scale). No color (white) indicates that no gene of V. cyclitrophicus ZF270 could be mapped to this element of the KEGG map. Boxes represent gene products, circles represent metabolites.
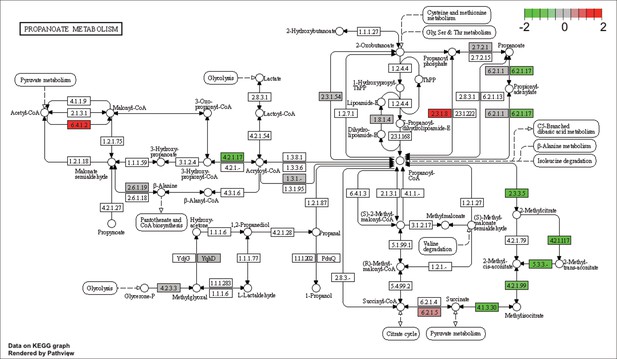
KEGG map of the significantly enriched KEGG pathway for propanoate metabolism.
The labeled KEGG maps were created with the R package pathview v.1.35.0 for each KEGG pathway that was identified as a significantly enriched gene set by GSEA (see Materials and methods). The color represents the log2 fold change of gene expression in V. cyclitrophicus ZF270 cells grown on digested alginate compared to alginate (green-grey-red color scale). No color (white) indicates that no gene of V. cyclitrophicus ZF270 could be mapped to this element of the KEGG map. Boxes represent gene products, circles represent metabolites.
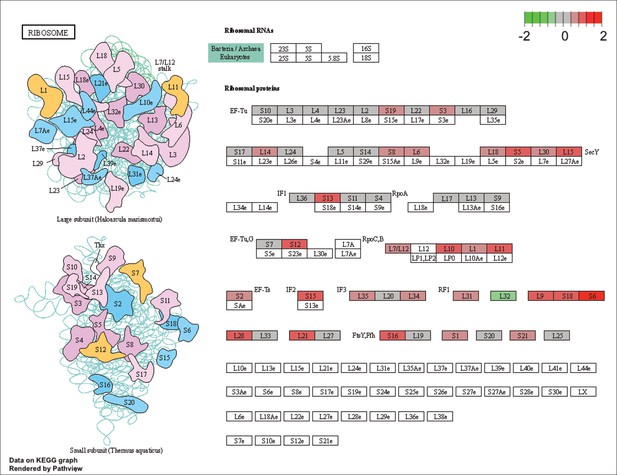
KEGG map of the significantly enriched KEGG pathway for ribosomal proteins.
The labeled KEGG maps were created with the R package pathview v.1.35.0 for each KEGG pathway that was identified as a significantly enriched gene set by GSEA (see Materials and methods). The color represents the log2 fold change of gene expression in V. cyclitrophicus ZF270 cells grown on digested alginate compared to alginate (green-grey-red color scale). No color (white) indicates that no gene of V. cyclitrophicus ZF270 could be mapped to this element of the KEGG map. Boxes represent gene products, circles represent metabolites.
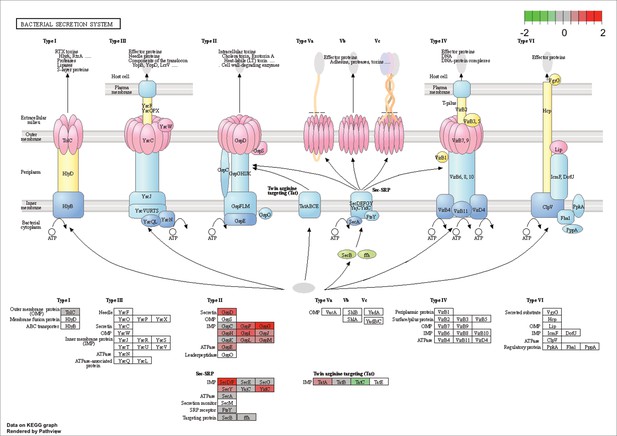
KEGG map of the significantly enriched KEGG pathway for bacterial secretion systems.
The labeled KEGG maps were created with the R package pathview v.1.35.0 for each KEGG pathway that was identified as a significantly enriched gene set by GSEA (see Materials and methods). The color represents the log2 fold change of gene expression in V. cyclitrophicus ZF270 cells grown on digested alginate compared to alginate (green-grey-red color scale). No color (white) indicates that no gene of V. cyclitrophicus ZF270 could be mapped to this element of the KEGG map. Boxes represent gene products, circles represent metabolites.
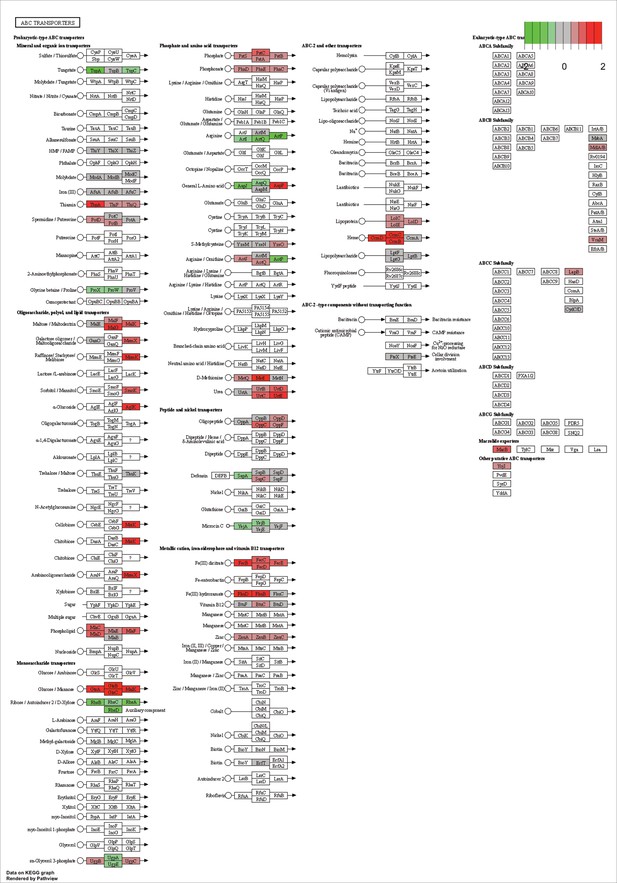
KEGG map of the significantly enriched KEGG pathway for ABC transporters.
The labeled KEGG maps were created with the R package pathview v.1.35.0 for each KEGG pathway that was identified as a significantly enriched gene set by GSEA (see Materials and methods). The color represents the log2 fold change of gene expression in V. cyclitrophicus ZF270 cells grown on digested alginate compared to alginate (green-grey-red color scale). No color (white) indicates that no gene of V. cyclitrophicus ZF270 could be mapped to this element of the KEGG map. Boxes represent gene products, circles represent metabolites.
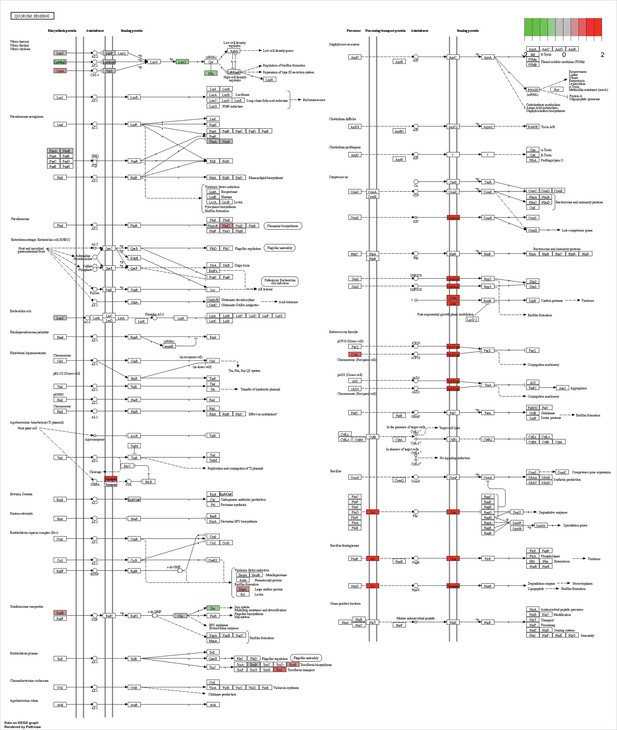
KEGG map of the significantly enriched KEGG pathway for quorum sensing.
The labeled KEGG maps were created with the R package pathview v.1.35.0 for each KEGG pathway that was identified as a significantly enriched gene set by GSEA (see Materials and methods). The color represents the log2 fold change of gene expression in V. cyclitrophicus ZF270 cells grown on digested alginate compared to alginate (green-grey-red color scale). No color (white) indicates that no gene of V. cyclitrophicus ZF270 could be mapped to this element of the KEGG map. Boxes represent gene products, circles represent metabolites.
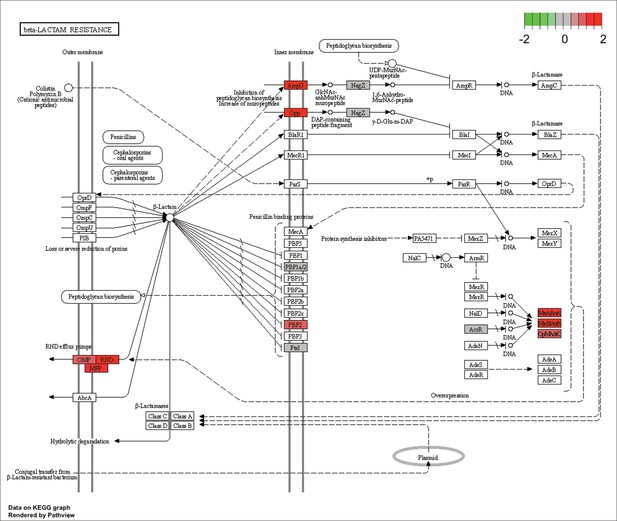
KEGG map of the significantly enriched KEGG pathway for beta-lactam resistance.
The labeled KEGG maps were created with the R package pathview v.1.35.0 for each KEGG pathway that was identified as a significantly enriched gene set by GSEA (see Materials and methods). The color represents the log2 fold change of gene expression in V. cyclitrophicus ZF270 cells grown on digested alginate compared to alginate (green-grey-red color scale). No color (white) indicates that no gene of V. cyclitrophicus ZF270 could be mapped to this element of the KEGG map. Boxes represent gene products, circles represent metabolites.
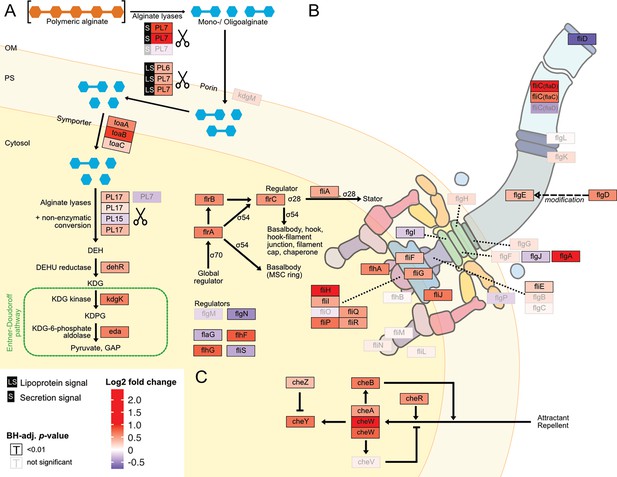
Digested alginate increases expression of genes involved in alginate degradation, uptake and catabolism, as well as flagellar assembly and chemotaxis.
Genome-wide differential expression analysis where the log2 fold changes of gene expression on digested alginate compared to alginate is shown for (A) alginate lyases (PL6, PL7, PL15, PL17, scissors symbol), transporters (porin kdgM, symporter toaB, symporter toaC), and metabolic enzymes shunting into the Entner-Doudoroff pathway (DEHU reductase DehR, kdgK, eda), (B) genes of the flagellar locus associated with flagellar assembly and (C) adjacent chemotaxis genes. Genes displayed in (B) and (C) are part of the KEGG pathways ‘Bacterial motility proteins’ and ‘Bacterial chemotaxis’. Differential expression analysis was performed to compute the Benjamini-Hochberg-adjusted Wald test p-value (‘BH-adj. p-value’, text color and box outline color) and log2 fold change (box fill color) for each gene (box). For better visibility, genes that exhibited a log2 fold gene expression change greater than 1 (i.e. doubling of expression) or less than –1 (i.e. halving of expression) are designated maximum intensity of red or blue, respectively. Genes with BH-adj. p-value smaller than 0.01 were considered significantly differentially expressed. In (A), the location of the gene products was based on Figure 1 of Wargacki et al., 2012 with the exception of the alginate lyases (PL6, PL7, PL15, PL17) which were placed based on their signal peptides (S: extracellular, LS: membrane-embedded, none: cytosolic). In (B) and (C) the gene location and depiction were based on the KEGG pathway ‘Flagellar assembly’ (map02040), ‘Bacterial chemotaxis’ (map02030), and Figure 3 of Rajagopala et al., 2007. Genes without known cellular location were omitted here but displayed in the genomic architecture in Figure 5—figure supplement 1. Arrow: activation; dashed arrow: modification; ‘flat’ arrow: inhibition; OM: outer membrane; PM: periplasm; IM: inner membrane; PL: polysaccharide lyase family; kdgM: oligogalacturonate-specific outer membrane porin; toaABC: oligoalginate symporter; DEH: 4-deoxy-L-erythro-5-hexoseulose uronic acid; dehR: DEH reductase; KDG: 2-keto-3-deoxy-gluconate; kdgK: KDG kinase; KDPG: 2-keto-3-deoxy-6-phosphogluconate; eda: KDG-6-phosphate aldolase; GAP: glyceraldehyde 3-phosphate; ED: Entner-Doudoroff; ns: not significant, that is BH-adj. p-value >0.01.
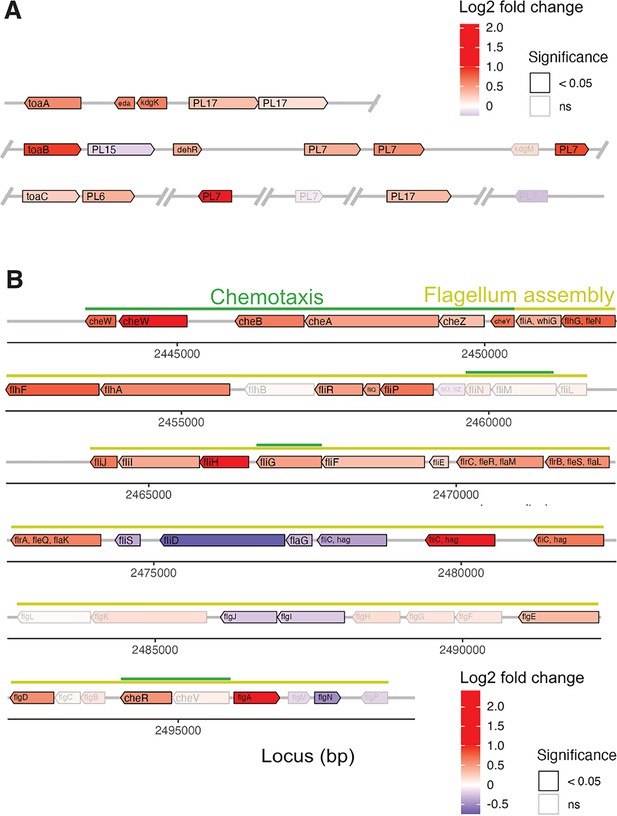
Genomic location and differential expression of genes encoding alginate catabolism and the flagellum locus.
Gene expression of V. cyclitrophicus ZF270 on digested alginate was compared to the gene expression on alginate using genome-wide differential expression analysis. The genomic location and the differential expression of genes encoding (A) alginate catabolism and (B) the flagellum locus are displayed. More specifically, the log2 fold changes in gene expression of (A) alginate lyases (PL6, PL7, PL15, PL17), transporters (porin kdgM, symporter toaB, symporter toaC), and metabolic enzymes shunting into the ED pathway (DEHU reductase DehR, kdgK, eda), and (B) genes of the flagellum assembly locus and adjacent chemotaxis genes based on KEGG pathway ‘Bacterial motility proteins’ and ‘Bacterial chemotaxis’ is displayed. Differential expression analysis was performed with DESeq2 v1.32.056 to compute the Benjamini-Hochberg-adjusted Wald test p-value (box color) and log2 fold change (box fill) for each gene. For better visibility, genes that exhibited a log2 fold gene expression change greater than 1 (i.e. doubling of expression) or less than –1 (i.e. halving of expression) are designated maximum intensity of red or blue, respectively. In A, the location of the gene products was based on Wargacki et al. (D’Souza et al., 2023b) with the exception of the alginate lyases (PL6, PL7, PL15, PL17) which were placed based on their signal peptide (S: extracellular, LS: membrane-anchored, none: cytosolic). OM: outer membrane; PM: periplasm; IM: inner membrane; PL: polysaccharide lyase family; kdgM: oligogalacturonate-specific outer membrane porin; toaABC: oligoalginate symporter; DEH: 4-deoxy-L-erythro-5-hexoseulose uronic acid; dehR: DEH reductase; KDG: 2-keto-3-deoxy-gluconate; kdgK: KDG kinase; KDPG: 2-keto-3-deoxy-6-phosphogluconate; eda: KDG-6-phosphate aldolase; GAP: glyceraldehyde 3-phosphate; ED: Entner-Doudoroff; ns: not significant.
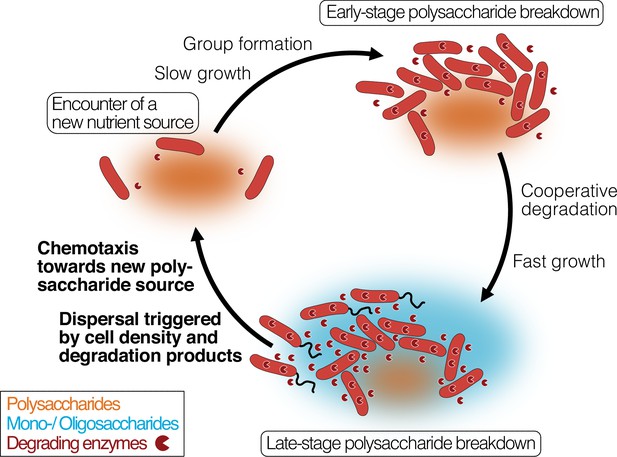
Bacterial growth and regulation on patches of polysaccharides.
By integrating our results with previous studies on cooperative growth on the same system, as well as results on dispersal cycles in other systems, we highlight where the specific results of this work add to this framework (bold font). When cells encounter polymer sources, the colonization of the nutrient hotspot may be aided by the basal exoenzyme production of ‘sentry’ enzymes (‘Encounter of a new carbon source’). This phase is succeeded by group formation, which enables cells to benefit from exoenzymes of neighboring cells and diffusing degradation products (‘Early-stage polysaccharide breakdown’). The following phase includes cooperative extracellular degradation of the polysaccharide source, further increasing the concentration of available degradation products. These degradation products trigger the overexpression of alginate degrading, importing, and catabolizing enzymes, ensuring swift polysaccharide degradation (‘Late-stage polysaccharide breakdown’). The increased pool of breakdown products also cues flagellar swimming in a subpopulation of cells and increases the expression of chemotaxis genes. Polymeric alginate acts as chemoattractant towards new polysaccharide sources. Cells and molecules are not drawn to scale. Dark red pie symbols: intracellular and extracellular polysaccharide-degrading enzymes; orange shading: a polymeric carbon source; blue shading: monomeric or oligomeric degradation products.
Additional files
-
Supplementary file 1
Differential gene expression of all genes of V. cyclitrophicus ZF270.
Genes of V. cyclitrophicus ZF270 were annotated by RASTtk. Differential expression analysis was performed on all genes with DESeq2 v1.32.0 Bassler et al., 1991 to compute the log2 fold change in gene expression for each gene and the corresponding p-value by Benjamini-Hochberg-adjusted Wald test. Geneid: gene identifier of genome annotation file; Chr: chromosome; Start: start of gene in base pairs; End: end of gene in base pairs; Strand: DNA strand on which gene is located; Length: length of gene; L1_raw to L6_raw: raw read count of replicate 1–6 on digested alginate; P1_raw to P6_raw: raw read count of replicate 1–6 on polymeric alginate; L1_DESeq to L6_DESeq: DESeq2-normalized read count of replicate 1–6 on digested alginate; P1_DESeq to P6_DESeq: DESeq2-normalized read count of replicate 1–6 on polymeric alginate; baseMean: baseMean value computed with DESeq2; log2FoldChange: log2 fold change value computed with DESeq2; lfcSE: shrunken (posterior) standard deviation computed with DESeq2; stat: Wald statistic computed with DESeq2, i.e. the log2 fold change divided by lfcSE, which is compared to a standard Normal distribution to generate a two-tailed p-value; pvalue: Wald test p-value computed with DESeq2; padj: Benjamini-Hochberg-adjusted Wald test p-value computed with DESeq2; RASTtk_Annotation: gene annotation by RASTtk; RASTtk_Ontology_term: ontology term by RASTtk; BlastKOALA_KO: KEGG Orthology by BlastKOALA; BlastKOALA_KO_Definition: KEGG Orthology definition by BlastKOALA; BlastKOALA_KO_Score: weighted sum of BLAST bit scores computed by BlastKOALA; KEGG_pathway: ID of the KEGG category C associated with the KEGG Orthology (BlastKOALA_KO), i.e., KEGG pathway ID or KEGG BRITE ID; KEGG_pathway_descr: Description of the KEGG category C; KEGG_CategB: ID of the KEGG category B associated with the KEGG Orthology (BlastKOALA_KO); KEGG_CategA: ID of the KEGG category A associated with the KEGG Orthology (BlastKOALA_KO). All KEGG categories were based on https://www.kegg.jp/kegg-bin/show_brite?ko00001.keg , Mar 18 2021.
- https://cdn.elifesciences.org/articles/93855/elife-93855-supp1-v1.xlsx
-
Supplementary file 2
Genome-wide pathway enrichment analysis.
Performed on all gene sets of KEGG category C (KEGG pathways and KEGG BRITE categories) by Gene Set Enrichment Analysis algorithm (GSEA) (Prouty et al., 2001). Method: fgsea() function and described filtering (see Materials and Methods); KEGG_hierarchy: ID of KEGG category C; KEGG_entry: KEGG pathway or KEGG BRITE category; Description: Description of the KEGG category; pval: enrichment p-value of GSEA; padj: BH-adjusted p-value of GSEA; log2err: the expected error for the standard deviation of the p-value logarithm, ES: enrichment score, same as in Broad GSEA implementation; NES: normalized enrichment score, normalized to mean enrichment of random samples of the same size; size: size of gene set after removing genes not present in the genome of V. cyclitrophicus ZF270; Genes_total_ZF270: number of genes of V. cyclitrophicus ZF270 within the gene set, counting gene duplicates.
- https://cdn.elifesciences.org/articles/93855/elife-93855-supp2-v1.xlsx
-
Supplementary file 3
Differential expression in genes of the valine, leucine and isoleucine biosynthesis (a subset of Supplementary file 1).
- https://cdn.elifesciences.org/articles/93855/elife-93855-supp3-v1.xlsx
-
Supplementary file 4
Differential expression in genes of the propanoate metabolism (a subset of Supplementary file 1).
- https://cdn.elifesciences.org/articles/93855/elife-93855-supp4-v1.xlsx
-
Supplementary file 5
Differential expression in genes encoding the ribosome (a subset of Supplementary file 1).
- https://cdn.elifesciences.org/articles/93855/elife-93855-supp5-v1.xlsx
-
Supplementary file 6
Differential expression in genes of the secretion system (a subset of Supplementary file 1).
- https://cdn.elifesciences.org/articles/93855/elife-93855-supp6-v1.xlsx
-
Supplementary file 7
Differential expression in genes of the bacterial secretion system (a subset of Supplementary file 1).
- https://cdn.elifesciences.org/articles/93855/elife-93855-supp7-v1.xlsx
-
Supplementary file 8
Differential expression in genes of the general secretion pathway (a subset of Supplementary file 1).
- https://cdn.elifesciences.org/articles/93855/elife-93855-supp8-v1.xlsx
-
Supplementary file 9
Differential expression in genes of enzymes with EC numbers (a subset of Supplementary file 1).
- https://cdn.elifesciences.org/articles/93855/elife-93855-supp9-v1.xlsx
-
Supplementary file 10
Differential expression in genes of transporters (a subset of Supplementary file 1).
- https://cdn.elifesciences.org/articles/93855/elife-93855-supp10-v1.xlsx
-
Supplementary file 11
Differential expression in genes of ABC transporters (a subset of Supplementary file 1).
- https://cdn.elifesciences.org/articles/93855/elife-93855-supp11-v1.xlsx
-
Supplementary file 12
Differential expression in genes of the bacterial motility proteins (a subset of Supplementary file 1).
- https://cdn.elifesciences.org/articles/93855/elife-93855-supp12-v1.xlsx
-
Supplementary file 13
Differential expression in genes associated with quorum sensing (a subset of Supplementary file 1).
- https://cdn.elifesciences.org/articles/93855/elife-93855-supp13-v1.xlsx
-
Supplementary file 14
Differential expression in genes of transcription factors (a subset of Supplementary file 1).
- https://cdn.elifesciences.org/articles/93855/elife-93855-supp14-v1.xlsx
-
Supplementary file 15
Differential expression in genes of beta-Lactam resistance (a subset of Supplementary file 1).
- https://cdn.elifesciences.org/articles/93855/elife-93855-supp15-v1.xlsx
-
Supplementary file 16
Differential expression of alginate lyases (PL6, PL7, PL15, PL17), transporters (porin kdgM, symporter toaB, symporter toaC), and metabolic enzymes shunting into the ED pathway (DEHU reductase DehR, kdgK, eda) (a subset of Supplementary file 1).
- https://cdn.elifesciences.org/articles/93855/elife-93855-supp16-v1.xlsx
-
Supplementary file 17
Differential expression of genes of the flagellum locus, comprising the cluster of genes that was part of the KEGG category of bacterial motility (a subset of Supplementary file 1).
- https://cdn.elifesciences.org/articles/93855/elife-93855-supp17-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/93855/elife-93855-mdarchecklist1-v1.pdf





