The BRAIN Initiative data-sharing ecosystem: Characteristics, challenges, benefits, and opportunities
Figures
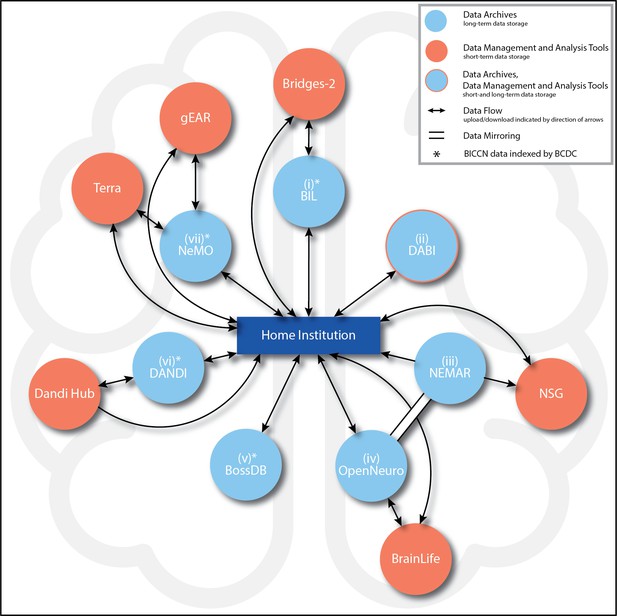
BRAIN Initiative data archive ecosystem connections.
Summary of the main BRAIN Initiative data archive ecosystem connections currently enabling data upload, access (ability to download), analysis (which includes short-term data storage), and mirroring (identical copies of data across archives) among entities. Potential paths of data flow are represented by arrows, with the directions of flow (upload, download, or both) indicated by the directions of the arrows. All seven archives (light blue circles) are intended for long-term data storage. For the archives marked with an asterisk (*), data generated by the BRAIN Initiative Cell Census Network (BICCN) also are indexed by the Brain Cell Data Center (BCDC) at the Allen Institute in Seattle, WA. Clockwise from top: (i) Brain Image Library (BIL): From the data producer’s Home institution (hereafter, ‘Home’) (dark blue rectangle), data can be uploaded to or downloaded from BIL. BIL data can be uploaded to, analyzed, and downloaded from the Bridges-2 supercomputer (orange circle). (ii) Data Archive for the BRAIN Initiative (DABI): From Home, data can be uploaded to or downloaded from DABI. (iii) NeuroElectroMagnetic data Archive and tools Resource (NEMAR): From Home, data can be downloaded from NEMAR. From NEMAR or Home, data also may be uploaded to the Neuroscience Gateway (NSG), although analyzed data may only be downloaded back to Home (and not back to NEMAR). All NEMAR data are mirrored (double solid line with no arrows) in OpenNeuro, and data also can only be uploaded to NEMAR through OpenNeuro. (iv) OpenNeuro: Data can be uploaded to or downloaded from OpenNeuro from Home. From OpenNeuro or Home, users also may upload and download data to BrainLife, a cloud-computing analysis environment. Through the data mirror, all OpenNeuro datasets are accessible on NEMAR. (v) Brain Observatory Storage Service and Database (BossDB): From Home, data can be uploaded to or downloaded from BossDB. (vi) Distributed Archives for Neurophysiology Data Integration (DANDI): From Home, data can be uploaded to and downloaded from DANDI. From Home, data analyzed in DANDI Hub also may be downloaded directly, but uploads to this analysis environment must always take place through DANDI. (vii) Neuroscience Multi-Omic Data Archive (NeMO): This archive is the primary repository for genomic, transcriptomic, and epigenetic data generated by the BICCN. From Home, data can be uploaded to or downloaded from NeMO. NeMO also streamlines user access to Terra and gEAR. By way of the BCDC, users also may link directly to Terra workspaces associated with NeMO projects. All descriptions current as of June 5, 2024.
Tables
BRAIN Initiative data archive ecosystem key statistics and related information.
Sources: For Host Institution, Initial Funding Date, Total NIH Support, Administering ICO, and Funding ICO, information was taken from the NIH RePORTER entry for the most recent award for each archive, as of June 6, 2024 (https://reporter.nih.gov/) Total NIH Support for each archive was taken from the History section of RePORTER. A separate line of NIH funding, totaling $1,707,134, with NIMH listed as the awardee, has supported the development of OpenNeuroPET. Other information in the table was derived from the International Neuroscience Coordinating Facility (INCF) Infrastructure Portfolio (https://www.incf.org/infrastructure-portfolio) and archive websites. Where an archive landing page or its data portal did not offer a public ‘datasets‘ (or ‘dandisets’, in the case of DANDI) count, we used an available alternative such as the number of ‘projects‘ for which data are available. *Data current as of June 6, 2024. ICO = NIH Institutes, Centers, and Offices.
| Archive | Host Institution | Initial Funding Date | Total NIH Support (USD)* | Administering ICO | Funding ICO | Data Types Hosted | Data Formats Supported | Publicly Available Datasets* |
|---|---|---|---|---|---|---|---|---|
| Brain Image Library (BIL) | Carnegie-Mellon University | 2017 | $6,767,625 | NIMH | NIMH | Confocal microscopy | DICOM or NIfTI; for derived data, any format as long as primary data are in the required format | 8418 |
| Brain Observatory Storage Service and Database (BossDB) | Johns Hopkins University | 2018 | $5,133,219 | NIMH | NIMH | Electron microscopy; x-ray microtomography | PNG; APNG; JPG; BMP; GIF; PSD | 50 |
| Data Archive for the BRAIN Initiative (DABI) | University of Southern California | 2018 | $6,160,745 | NIMH | NINDS | Focus on invasive neurophysiology; houses all imaging and brain signal data | EDF, Biosemi (.bdf), BrainVision (.eeg,.vhdr,.vmrk), EEGLAB (.set,.fdt), Blackrock NeuroPort (.nev,.nsX), Intan (.rhd,.rhs), MATLAB files (.mat,.m); iEEG-BIDS, NWB, or DABI | 110 |
| Distributed Archives for Neurophysiology Data Integration (DANDI) | Massachusetts Institute of Technology | 2019 | $8,309,010 | NIMH | NIMH | Cellular neurophysiology, neuroimaging, and microscopy | BIDS; NWB | 640 |
| NeuroElectroMagnetic data Archive and tools Resource (NEMAR) | University of California, San Diego | 2019 | $4,464,874 | NIMH | NIMH | EEG, MEG | BIDS | 297 |
| Neuroscience Multi-Omic Data Archive (NeMO) | University of Maryland, Baltimore | 2017 | $9,347,683 | NIMH | NIMH | Multi-omics | FASTQ; TSV; BAM; BIGWIG; MEX; LIST; BED; QBED; CSV; H5; BIGBED; LOOM; MTX; ASPX | 49 |
| OpenNeuro | Stanford University | 2018 | $7,254,848 | NIMH | NIMH | MRI, PET, MEG, EEG, iEEG | BIDS | 1076 |





