Expanding the Drosophila toolkit for dual control of gene expression
Figures
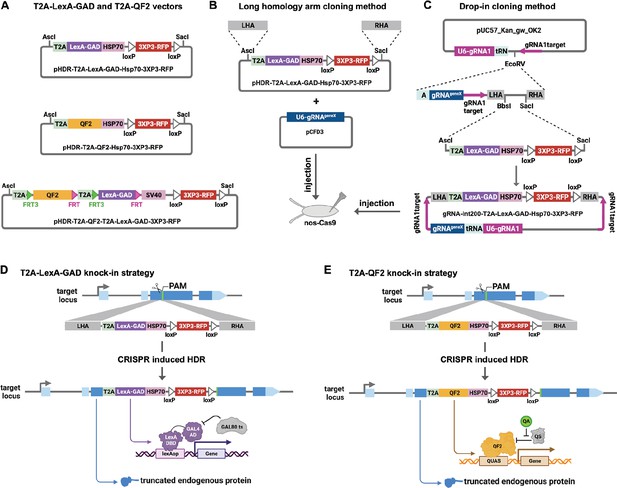
Strategy for CRISPR knock-in of T2A-LexA-GAD and T2A-QF2.
(A) Donor vectors for knock-in. pHDR-T2A-LexA-GAD-Hsp70-3xP3-RFP and pHDR-T2A-QF2-Hsp70-3xP3-RFP contain T2A-LexA-GAD or QF2 transcriptional activators followed by HSP70 terminators. pHDR-T2A-QF2-T2A-LexA-GAD-3xP3-RFP contains both activators flanked by FRT sites followed by SV40 terminator. All the vectors contain a 3XP3-RFP transformation marker flanked by loxP sites. (B) Long homology arm cloning method. ~1000 bp homology arms are amplified from genomic DNA and inserted into the AscI and SacI sites by Gibson assembly. A separate target-gene-specific guide RNA is cloned into U6 promoter expression vector such as pCFD3. (C) Drop-in cloning method. Based on the gRNA-int200 method previously described (Kanca et al., 2022). A company synthesizes and clones a DNA fragment into the pUC57_Kan_gw_OK2 vector. The resulting plasmid contains the following elements: (1) two guide RNAs under the control of a U6 promoter, one (gRNA1) targeting the vector (pink arrows) to linearize the homology donor in vivo, and another (gRNAgeneX) targeting the gene of interest; (2) a tRNA sequence to allow liberation of the individual guides by the endogenous tRNA processing machinery; (3) 200 bp short homology arms; and (4) BbsI and SacI cloning sites. The AscI/SacI T2A-LexA-GAD, T2A-QF, or T2A-LexA-GAD-T2A-QF fragments can then be ligated in a single directional cloning step into the BbsI/SacI sites to produce the donor plasmid. (D) T2A-LexA-GAD and (E) T2A-QF2 knock-in strategy. CRISPR-based HDR causes integration of the T2A-LexA-GAD or T2A-QF2 in the most 5’ coding exon common to all or most isoforms, resulting in expression of the activators under control of the endogenous gene regulatory region. The knock-in also produces a truncated endogenous protein and thus a strong loss-of-function allele.
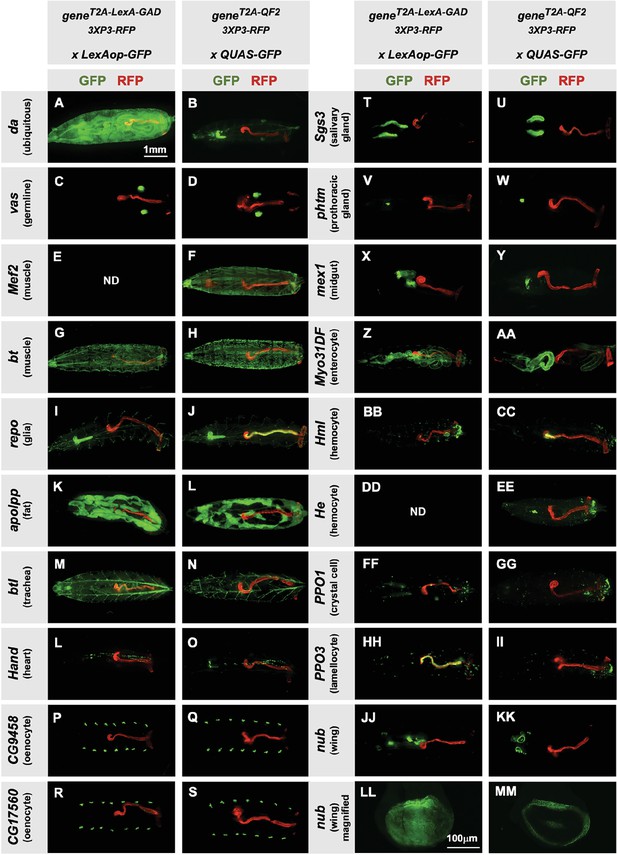
Tissue specificity of T2A-LexA-GAD and T2A-QF2 knock-ins.
(A–KK) T2A-LexA-GAD knock-in lines crossed to a LexAop-GFP reporter and T2A-QF2 knock-in lines crossed to a QUAS-GFP reporter. Panels show third instar larva. GFP shows the driver line expression pattern. RFP shows the 3XP3 transformation marker, which labels the posterior gut and anal pads of the larva. Gene names and tissues are on the left. We failed to obtain LexA-GAD knock-ins for Mef2 (E) and He (DD). (LL–MM) Third instar imaginal disc from the insertions in the nubbin (nub) gene. Note that most of the lines are highly tissue-specific and are comparable between the LexA-GAD and QF2 knock-ins. Insertions in the daughterless gene (da) and nub are an exception, as the T2A-LexA-GAD, but not the T2A-QF2, gives the expected expression pattern. Insertions in the gut-specific genes mex1 (X–Y) and Myo31Df (Z–AA) also differed between the LexA-GAD and QF2 drivers.
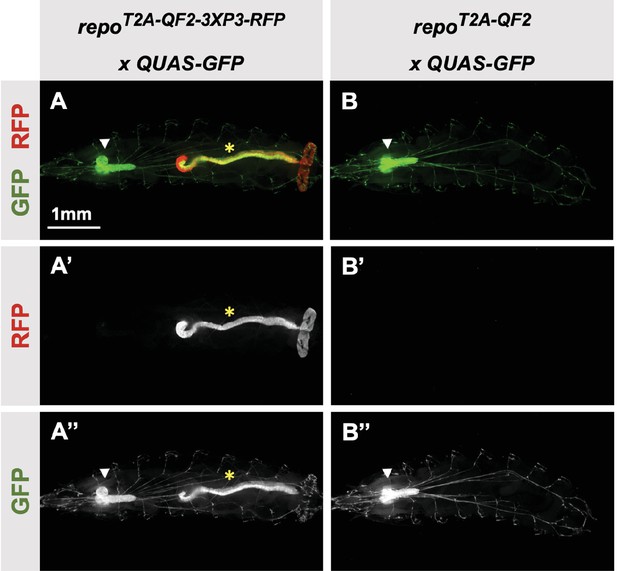
3XP3-RFP can cause misexpression of T2A-LexA-GAD or T2A-QF2.
(A) T2A-QF2-3XP3-RFP in the repo gene crossed to a QUAS-GFP reporter. In third instar larva, the reporter is expressed in the expected glial cells, but also misexpressed in gut and anal pad (yellow asterisk). (B) T2A-QF2 in the repo gene with the 3XP3-RFP removed by Cre-Lox recombination, crossed to a QUAS-GFP reporter. Removal of 3XP3-RFP eliminated gut and anal pad misexpression and did not affect glial cell expression (white arrowheads).
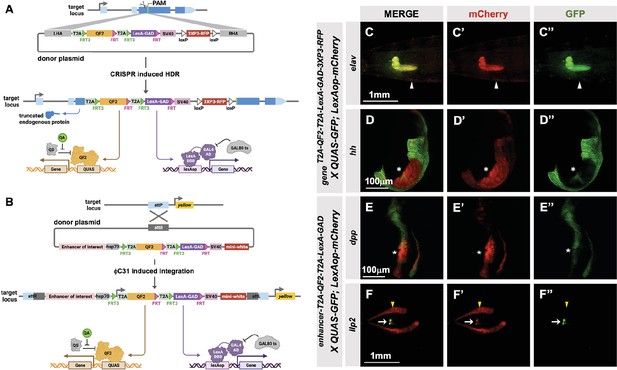
T2A-QF2-T2A-LexA-GAD double driver lines.
(A) CRISPR-based HDR strategy for integration of the T2A-QF2-T2A-LexA-GAD-3XP3 in the most 5’ coding exon common to all or most isoforms, resulting in expression of both activators under control of the endogenous gene regulatory region. The knock-in also produces a truncated endogenous protein and thus a strong loss-of-function allele. If desired, one of the two coding regions can then be excised with Flp, resulting in flies that express only QF2 or LexA-GAD. (B) Alternative strategy allows gene enhancers to be cloned upstream of of T2A-QF2-T2A-LexA-GAD. The vector backbone includes an attB site for phiC31 insertion into attP flies. (C–D) T2A-QF2-T2A-LexA-GAD knock-ins crossed to a QUAS-GFP+LexAop-mCherry double reporter line. (C) The elavT2A-QF2-T2A-LexA-GAD knock-in drives both QUAS-GFP and LexAop-mCherry in the larval brain. There is some leakiness of mCherry in the body wall muscle (arrowheads). (D) The hhT2A-QF2-T2A-LexA-GAD knock-in drives both QUAS-GFP and LexAop-mCherry in the posterior of the wing imaginal disc. GFP expression is much less than mCherry in the wing pouch (asterisks). (E–F) Enhancer-T2A-QF2-T2A-LexA-GAD lines crossed to a QUAS-GFP+LexAop-mCherry double reporter line. (E) The dpp-blk enhancer-T2A-QF2-T2A-LexA-GAD line drives both QUAS-GFP and LexAop-mCherry along the anterior/posterior boundary of the wing imaginal disc. GFP expression is much less than mCherry in the wing pouch (stars). (F) The Ilp2 enhancer-T2A-QF2-T2A-LexA-GAD line drives both QUAS-GFP and LexAop-mCherry in the insulin-producing cells of the larval brain (arrows). The fat body mCherry expression (yellow arrowhead) is from leakiness of the reporter stock and does not indicate LexA-GAD activity.
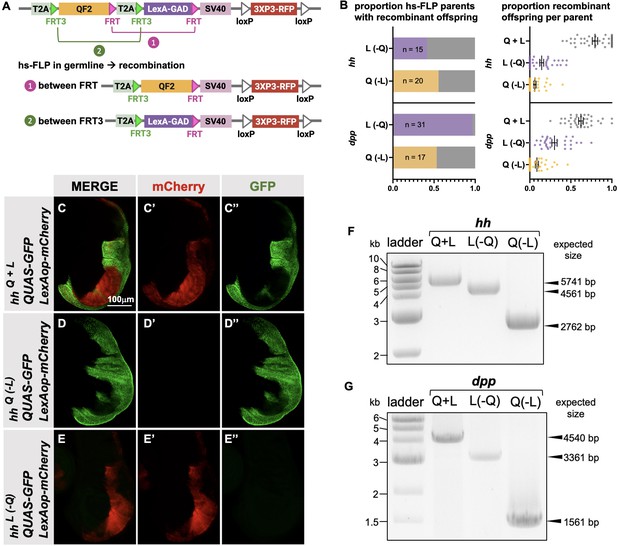
Generation of single drivers from T2A-QF2-T2A-LexA-GAD knock-ins by hs-FLP.
(A) FLP/FRT recombination scheme. Flies containing both hs-FLP and a T2A-QF2-T2A-LexA-GAD knock-in are heat shocked during larval development to induce one of two mutually exclusive recombination events in their germline between either FRT or FRT3. (B) Heat shock of hsFLP; hhT2A-QF2-T2A-LexA-GAD and hsFLP; dppT2A-QF2-T2A-LexA-GAD flies produces frequent recombinants, both T2A-QF2 and T2A-LexA-GAD. The bar graph shows the proportion of heat-shocked animals that produced at least one recombinant offspring. The dot plot shows the proportion of recombinant offspring per heat-shocked parent. Mean ± SD is indicated. (C–D) Validation of individual hhT2A-QF2 and hhT2A-LexA-GAD derivatives by immunofluorescence. All panels show third instar larval wing discs dissected from potential hhT2A-QF2-T2A-LexA-GAD recombinants crossed to a QUAS-GFP+LexAop-mCherry reporter line. (C) Wing disc from non-recombinant hhT2A-QF2-T2A-LexA-GAD showing expression of both GFP and mCherry in the posterior of the wing disc. Note, this is the same image as shown in Figure 4D. (D) Wing disc from recombinant hhT2A-QF2 showing expression of GFP but not mCherry in the posterior of the wing disc. (E) Wing disc from recombinant hhT2A-LexA-GAD showing expression of mCherry but not GFP in the posterior of the wing disc. Validation of (F) hhT2A-QF2 and hhT2A-LexA-GAD derivatives and (G) dppT2A-QF2 and dppT2A-LexA-GAD derivatives by PCR from genomic DNA from individual flies. In all panels, for brevity, T2A-QF2-T2A-LexAop, T2A-QF2, and T2A-LexAop, are notated as Q+L, Q(-L), and L(-Q), respectively.
-
Figure 5—source data 1
Original file for the DNA gel analysis in Figure 5F and G.
- https://cdn.elifesciences.org/articles/94073/elife-94073-fig5-data1-v1.zip
-
Figure 5—source data 2
Original file for the DNA gel analysis with boxes indicating the portions cropped to produce the image for Figure 5F and G.
- https://cdn.elifesciences.org/articles/94073/elife-94073-fig5-data2-v1.zip
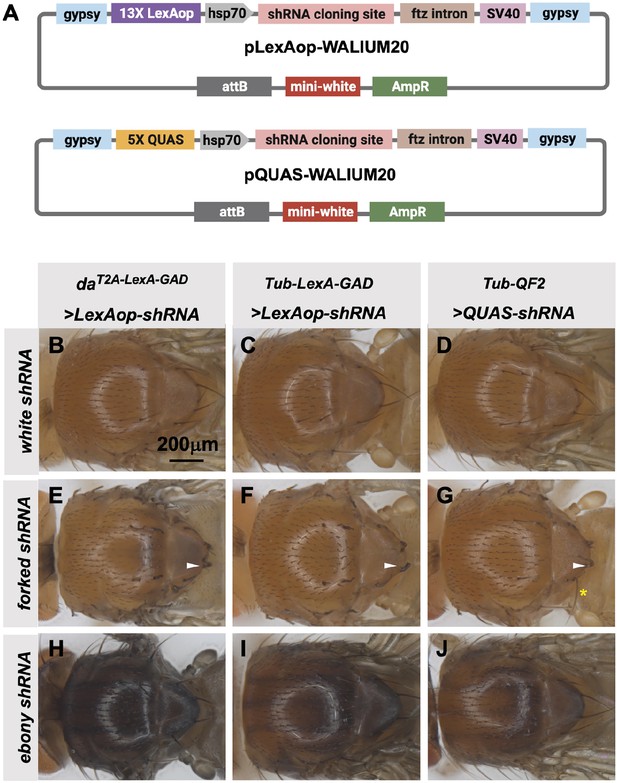
Transgenic RNAi Project (TRiP) LexAop and QUAS shRNA vectors produce effective gene knockdown.
(A) shRNAs for knockdown or genes for overexpression were cloned into pLexAop-WALIUM20 and pQUAS-WALIUM20, derived from the TRiP WALIUM20 vector. (B–J) Dorsal view of adult fly thoraces resulting from crosses of LexAop or QUAS shRNAs to daT2A-LexA-GAD (generated in this study), Tub-LexA-GAD (Bloomington Drosophila Stock Center [BDSC] 66686), or Tub-QF2 (BDSC 51958). (B–C) white shRNA control produced no thoracic phenotypes in any of the crosses. (E–G) forked shRNA produced a forked bristles phenotype (white arrowheads). Note that some bristles retain a more elongated wild-type morphology with the Tub-QF2-driven forked knockdown (G, yellow asterisk). (H–J) ebony shRNA produced a darkened cuticle phenotype. The daT2A-LexA-GAD driver produced the strongest phenotype (compare panel H to I and J).
Tables
T2A-LexA-GAD and T2A-QF2 knock-in lines.
| Target tissue | Target gene (name) | QF2 | This paper | 3XP3 removed | Others at BDSC | LexA-GAD | This paper | 3XP3 removed | Others at BDSC |
|---|---|---|---|---|---|---|---|---|---|
| Ubiquitous | da | Yes | Yes | Yes | No | Yes | Yes | Yes | No |
| Germline | vas* | Yes | Yes | No | No | Yes | Yes | No | No |
| Muscle | Mef2 | Yes | Yes | Yes | 66469 | Yes | No | Yes | 97530 |
| Muscle | bt* | Yes | Yes | No | No | Yes | Yes | No | No |
| Pan-neuronal | elav† | Yes | Yes | Yes | 66466 | Yes | Yes | Yes | No |
| Glia | repo | Yes | Yes | No | 66477 | Yes | Yes | Yes | 97535 |
| Insulin-producing cells | Ilp2 (regulatory region)† | Yes | Yes | N/A | No | Yes | Yes | N/A | No |
| Fat | apolpp | Yes | Yes | Yes | No | Yes | Yes | Yes | No |
| Trachea | btl | Yes | Yes | Yes | No | Yes | Yes | Yes | No |
| Heart | Hand* | Yes | Yes | Yes | No | Yes | Yes | Yes | No |
| Enocyte | CG9458* | Yes | Yes | No | No | Yes | Yes | Yes | No |
| Enocyte | CG17560* | Yes | Yes | No | No | Yes | Yes | No | No |
| Salivary gland | Sgs3* | Yes | Yes | No | No | Yes | Yes | Yes | No |
| Prothoracic gland | phtm | Yes | Yes | Yes | No | Yes | Yes | Yes | No |
| Midgut | mex1 | Yes | Yes | Yes | No | Yes | Yes | Yes | No |
| Adult midgut enterocyte | Myo31DF | Yes | Yes | Yes | No | Yes | Yes | Yes | No |
| Hemocyte | Hml | Yes | Yes | Yes | 66468 | Yes | Yes | Yes | No |
| Hemocyte | He | Yes | Yes | Yes | No | No | No | N/A | No |
| Crystal cell | PPO1 | Yes | Yes | Yes | No | Yes | Yes | Yes | No |
| Lamellocyte | PPO3 | Yes | Yes | Yes | No | Yes | Yes | Yes | No |
| Posterior segment | hh† | Yes | Yes | Yes | No | Yes | Yes | Yes | 97536 |
| Wing pouch/hinge | nub | Yes | Yes | Yes | No | Yes | Yes | Yes | No |
| Imaginal disc A/P boundary | dpp (regulatory region)† | Yes | Yes | N/A | No | Yes | Yes | N/A | No |
-
*
Constructs were cloned using the drop-in method. All others were cloned by PCR of long homology arms.
-
†
Constructs were cloned into double driver vectors.
Additional files
-
Supplementary file 1
Table listing the gene names and antisense sequence of 96 constructs cloned and injected to produce a set of transgenic LexAop-shRNA fly lines.
- https://cdn.elifesciences.org/articles/94073/elife-94073-supp1-v1.xlsx
-
Supplementary file 2
Table listing the gene names, guide sequences, and primers for amplification of long homology arms for T2A-QF2 and T2A-LexA-GAD constructs.
- https://cdn.elifesciences.org/articles/94073/elife-94073-supp2-v1.xlsx
-
Supplementary file 3
Sequences of DNA fragments synthesized into the pUC57 Kan_gw_OK2 vector backbone for subsequent drop-in cloning.
- https://cdn.elifesciences.org/articles/94073/elife-94073-supp3-v1.xlsx
-
Supplementary file 4
Sequences of DNA fragments used to build pHDR-T2A-QF2-T2A-LexA-GAD and pMCS-T2A-QF2-T2A-LexA-GAD-WALIUM20.
FRT3 sequences are shown in blue and FRTwt sequences are shown in red.
- https://cdn.elifesciences.org/articles/94073/elife-94073-supp4-v1.xlsx
-
Supplementary file 5
Primers and guide sequences for amplification of long homology arms for pHDR-T2A-QF2-T2A-LexA-GAD-3XP3-RFP constructs.
- https://cdn.elifesciences.org/articles/94073/elife-94073-supp5-v1.xlsx
-
Supplementary file 6
Primers used to amplify large enhancers for cloning into pMCS-T2A-QF2-T2A-LexA-GAD-WALIUM20.
The dpp-blk primers have NotI and EcoRI sites for cloning into the MCS, and the Dilp2-enh primers have PacI and EcoRI sites.
- https://cdn.elifesciences.org/articles/94073/elife-94073-supp6-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/94073/elife-94073-mdarchecklist1-v1.pdf





