CXCR3-expressing myeloid cells recruited to the hypothalamus protect against diet-induced body mass gain and metabolic dysfunction
Figures
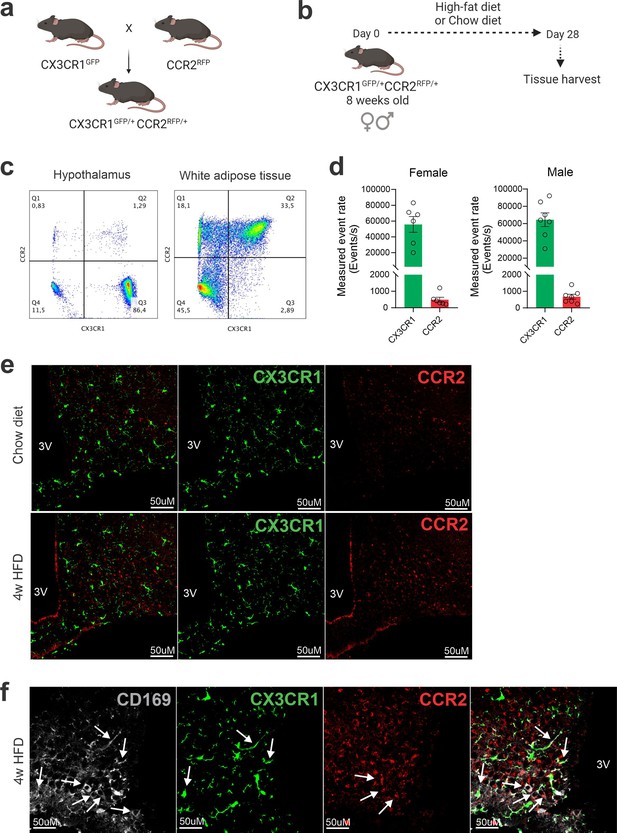
CCR2-positive cells infiltrate the hypothalamus of mice fed a high-fat diet (HFD).
(a) CX3CR1GFP/+CCR2RFP/+ dual-reporter mutant mice generation. (b) Schematic representation of the experimental protocol for analysis of HFD-induced CCR2 peripheral-cell chemotaxis toward the hypothalamus. (c) Flow cytometry analysis of CX3CR1GFP+ and CCR2RFP+ cells in the white adipose tissue and in the hypothalamus of HFD-fed mice. (d) Measured event rate detected by flow cytometer of CX3CR1GFP+ and CCR2RFP+ cells isolated from the hypothalamus of HFD-fed male and female mice. (e) Coronal brain sections of mediobasal hypothalamus (MBH) from chow- and 4 weeks HFD-fed mice CX3CR1GFP/+CCR2RFP/+. (f) Coronal brain sections of MBH from 4 weeks HFD-fed mice CX3CR1GFP/+CCR2RFP/+ immunostained for CD169 (Sialoadhesin). White arrows indicate overlap between CD169-positive cells with CX3CR-positive cells or with CCR2-positive cells. 3V = third ventricle, scale bar = 50 μm.
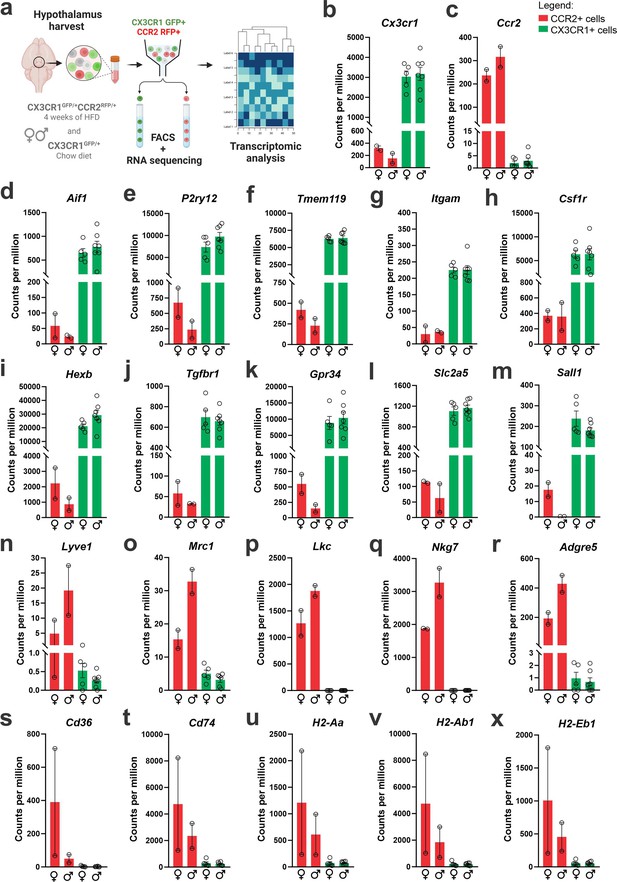
CX3CR1-positive resident microglia and CCR2-positive recruited myeloid cells sorted from the hypothalamus of high-fat diet (HFD)-fed mice express classical markers of microglia and other immune cells.
(a) Schematic representation of the experimental protocol for sorting and sequencing CX3CR1 and CCR2 cells from the hypothalamus of chow- and HFD-fed mice. (b) Cx3cr1 gene expression and (c) Ccr2 gene expression of CX3CR1 and CCR2 cells sorted from the hypothalamus of HFD-fed mice. Analysis of (d–m) classical microglial markers, and (n–x) bone marrow-derived immune cell markers in the transcriptome of CX3CR1 and CCR2 cells sorted from the hypothalamus of HFD-fed mice. To perform RNA-sequencing (RNA-seq) we have employed a total of 200 mice of each sex fed on HFD and 100 mice of each sex fed on chow diet. They were divided into five independent experiments. To get the total RNA amount from CCR2-positive cells in the hypothalamus of HFD-fed mice that was enough for library construction and RNA-seq, CCR2 samples were pooled together in three samples, but only two samples could be sequenced due to the final RNA integrity and amount.
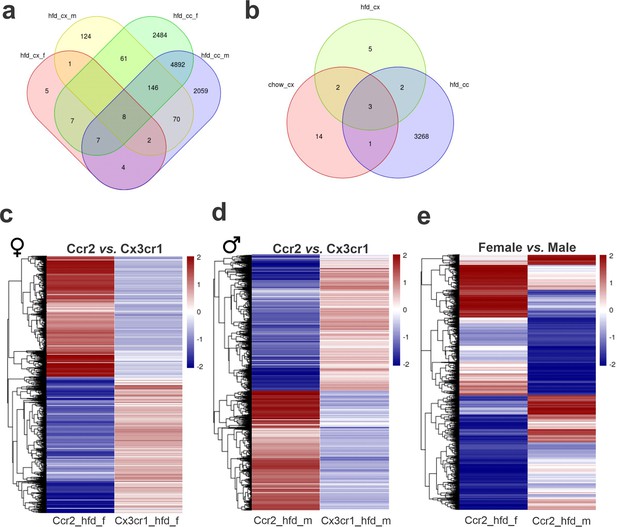
Differential gene expression (DGE) analysis of CX3CR1-positive resident microglia and CCR2-positive recruited myeloid cells sorted from the hypothalamus of high-fat diet (HFD)-fed mice show an enormous difference in their transcriptomic signature.
(a, b) Venn diagram showing the number of DGEs for chow and HFD diet comparison and sex comparison, respectively. (c) Heatmap of up- and downregulated DGEs when comparing CX3CR1 and CCR2 cells from HFD-fed female mice. (d) Heatmap of up- and downregulated DGEs when comparing CX3CR1 and CCR2 cells from HFD-fed male mice. (e) Heatmap of up- and downregulated DGEs when comparing CCR2 recruited myeloid cells from HFD-fed male and female mice. Legend: hfd_cx_f = Cx3cr1_hfd_female vs. Cx3cr1_chow_female; hfd_cx_m = Cx3cr1_hfd_male vs. Cx3cr1_chow_male; chow_cx = Cx3cr1_chow_male vs. Cx3cr1_chow_female; hfd_cx = Cx3cr1_hfd_male vs. Cx3cr1_hfd_female; hfd_cc_f = Cx3cr1_hfd_female vs. Ccr2_hfd_female; hfd_cc_m = Cx3cr1_hfd_male vs. Ccr2_hfd_male; hfd_cc = Ccr2_hfd_male vs. Ccr2_hfd_female.
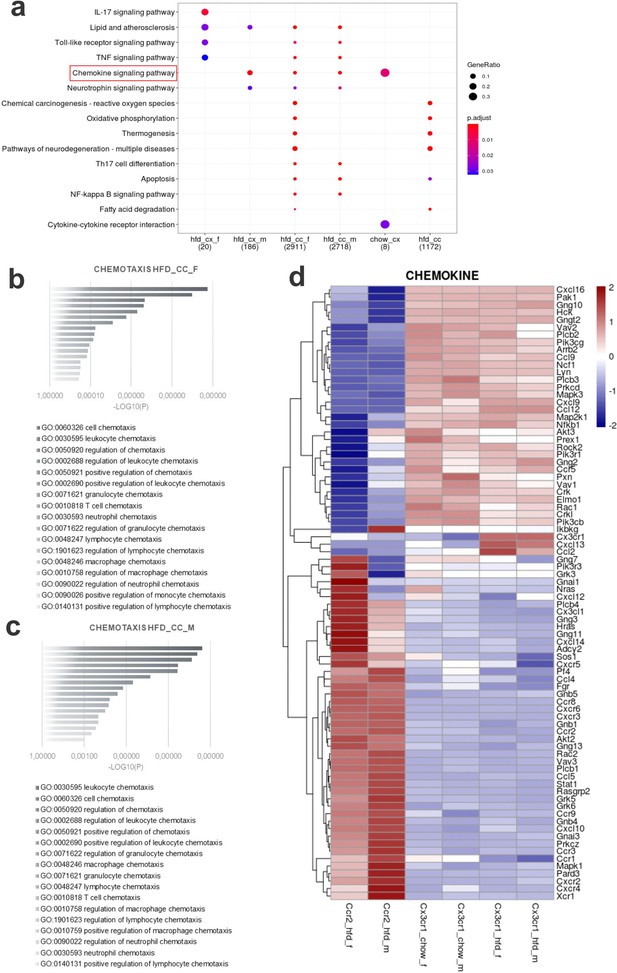
Various differential gene expression (DGE) found in CCR2+ infiltrating cells from the hypothalamus of high-fat diet (HFD)-fed mice belong to chemotaxis pathways.
(a) Kioto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis shows the distribution of DGEs in distinct metabolic pathways. (b, c) Ontology analysis for DGEs related to chemotaxis from CCR2-positive cells sorted from the hypothalamus of HFD-fed female and male mice, respectively. (d) Heatmap of up- and downregulated DGEs related to chemotaxis when comparing CX3CR1-positive microglia and CCR2-positive recruited myeloid cells from HFD-fed male and female mice.
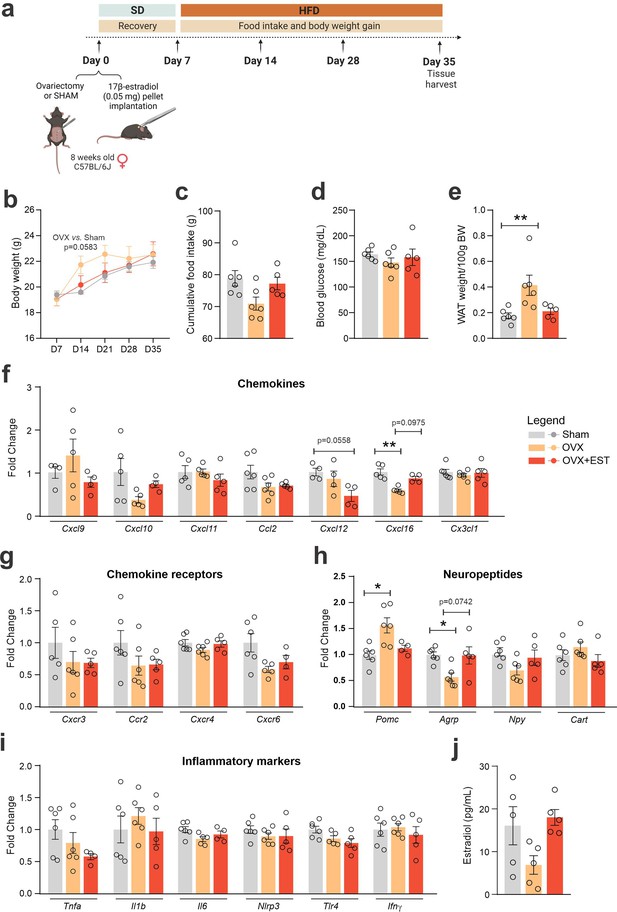
Ovariectomy and ovariectomy with estradiol replacement modulate hypothalamic chemokine and neuropeptides.
(a) Experimental design of the ovariectomy and estradiol replacement protocol performed in C57BL/6J female mice; (b) body mass gain from Day 7 to 35 of the experimental protocol. (c) Total food intake during the experimental period. (d) Fasting blood glucose levels and (e) retroperitoneal white adipose tissue retroperitoneal depot weight at Day 35. Hypothalamic mRNA levels of (f) chemokines; (g) chemokine receptors; (h) neuropeptides; (i) inflammatory genes; (j) blood estradiol. Data are expressed as mean ± SEM of 4–6 mice/group. One- and two-way ANOVA followed by Sidak’s post hoc test and Mann–Whitney test were used for statistical analyses. *p < 0.05 and **p < 0.01, in comparison with the Sham group.
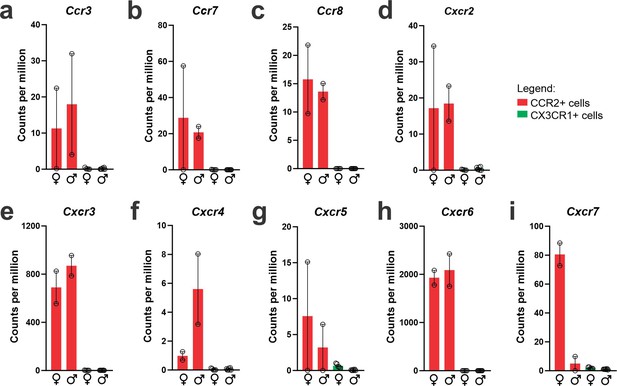
CCR2-positive recruited myeloid cells from the hypothalamus of high-fat diet (HFD)-fed mice express a broad range of chemokine receptors.
(a–i) Chemokine receptors gene expression in the transcriptome of CX3CR1- and CCR2-positive cells sorted from the hypothalamus of HFD-fed mice.
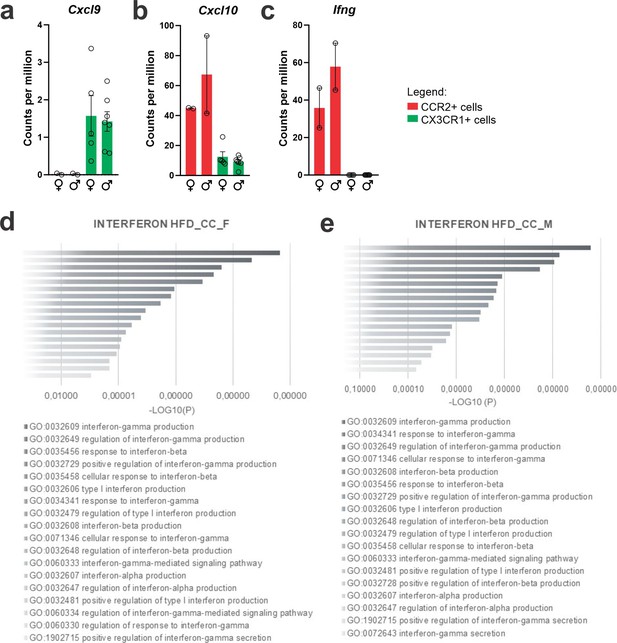
CXCL10/interferon γ-induced protein 10 kDa (IP-10) is highly expressed in CCR2-poisitve recruited myeloid cells from the hypothalamus of high-fat diet (HFD)-fed mice.
(a–c) Cxcl9, Cxcl10, and Ifng gene expression in the transcriptome of CX3CR1- and CCR2-positive cells sorted from the hypothalamus of HFD-fed mice. (d, e) Ontology analysis for DGEs related to interferon signaling pathways from CCR2-positive cells sorted from the hypothalamus of HFD-fed female and male mice, respectively.
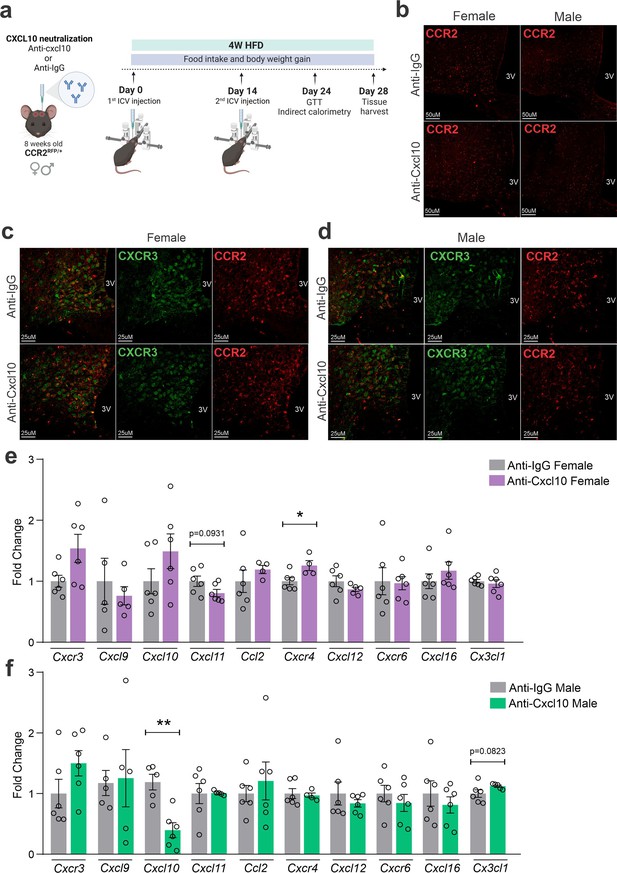
CXCL10 neutralization has a mild impact on reducing CCR2- and CXCR3-positive cell chemotaxis toward the hypothalamus of high-fat diet (HFD)-fed mice.
(a) Schematic representation of the experimental protocol for CXCL10 central neutralization. (b) Coronal brain sections from 4 weeks HFD-fed CCR2RFP/+ mice showing the CCR2-positive cells distribution in the hypothalamic parenchyma upon CXCL10 central neutralization. 3V = third ventricle, scale bars = 50 μm. (c) Coronal brain sections from 4 weeks HFD-fed female CCR2RFP/+ mice immunostained for CXCR3 upon CXCL10 central neutralization. (d) Coronal brain sections from 4 weeks HFD-fed male CCR2RFP/+ mice immunostained for CXCR3 upon CXCL10 central neutralization. 3V = third ventricle, scale bars = 25 μm. (e, f) Hypothalamic mRNA levels of several chemokine receptors and chemokines in HFD-fed female (light gray and purple bars) and male (light gray and green bars) CCR2RFP/+ mice upon CXCL10 central neutralization. For qualitative confocal image analysis, we have used 3 samples per group. For real-time quantitavive polymerase chain reaction (RT-qPCR) of the hypothalamus, we have used 5–6 samples per group. Two-tailed Mann–Whitney tests were used for statistical analyses. *p < 0.05 and **p < 0.01 in comparison with respective Anti-IgG-treated groups.
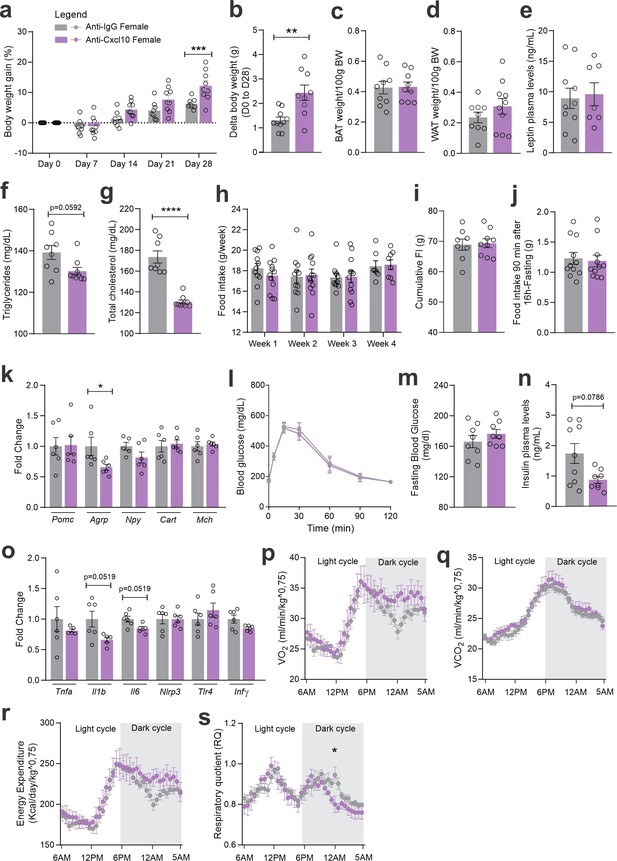
CXCL10 central neutralization in high-fat diet (HFD)-fed female mice.
(a) Percentual of body weight gain from Day 0 to 28 of the experimental protocol. (b) Delta body weight during the experimental period. (c) Brown adipose tissue weight and (d) white adipose tissue (retroperitoneal depot) weight at Day 28. (e) Leptin, (f) triglycerides, and (g) total cholesterol plasma levels at Day 28. (h) Weekly food intake measurement during the experimental period. (i) Cumulative food intake during the experimental period. (j) 90-min food intake measurement after 16 hr of fasting. (k) Hypothalamic mRNA levels of neuropeptides involved in food intake control. (l) Intraperitoneal glucose tolerance test on Day 24. (m) 6-hr fasting blood glucose levels. (n) Insulin plasma levels at Day 28. (o) Hypothalamic mRNA levels of inflammatory genes. (p) O2 consumption; (q) CO2 production; (r) energy expenditure; (s) respiratory quotient at Day 24. Data were expressed as mean ± SEM of 8–10 mice/group (in two independent experiments). To perform quantitative reverse transcription-polymerase chain reaction (qRT-PCR) we have used 6 mice/group. To perform biochemical analysis in plasma we have used 8–10 mice/group. To perform ipGTT we have used 4 mice/group. To perform indirect calorimetry, we have used 4 mice/group. Two-way ANOVA followed by Sidak’s post hoc test and Mann–Whitney test were used for statistical analyses. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 in comparison with IgG-treated group.
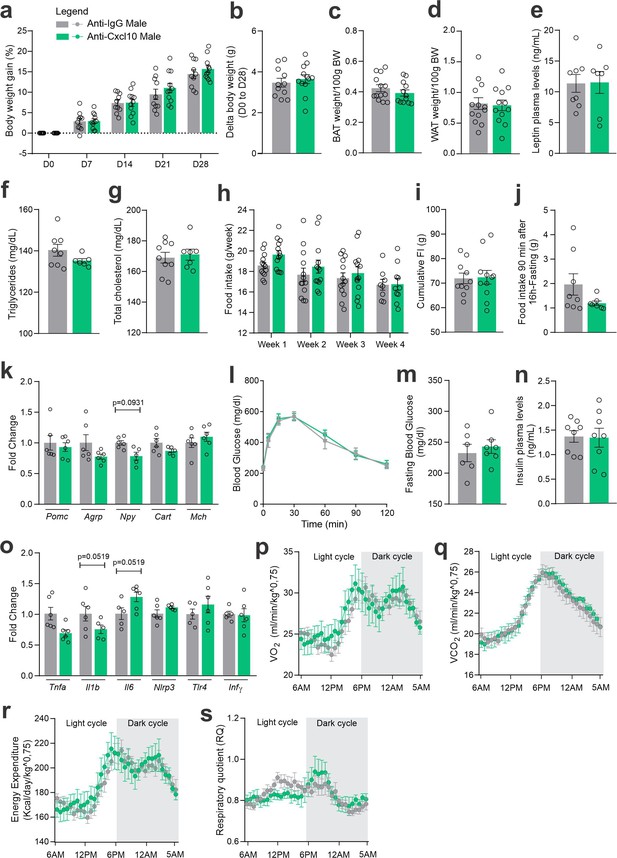
CXCL10 central neutralization in high-fat diet (HFD)-fed male mice.
(a) Percentual of body weight gain from Day 0 to 28 of the experimental protocol. (b) Delta body weight during the experimental period. (c) Brown adipose tissue weight and (d) white adipose tissue (retroperitoneal depot) weight at Day 28. (e) Leptin, (f) triglycerides, and (g) total cholesterol plasma levels at Day 28. (h) Weekly food intake measurement during the experimental period. (i) Cumulative food intake during the experimental period. (j) 90-min food intake measurement after 16 hr of fasting. (k) Hypothalamic mRNA levels of neuropeptides involved in food intake control. (l) Intraperitoneal glucose tolerance test at Day 24. (m) 6-hr fasting blood glucose levels. (n) Insulin plasma levels at Day 28. (o) Hypothalamic mRNA levels of inflammatory genes. (p) O2 consumption; (q) CO2 production; (r) energy expenditure; (s) respiratory quotient at Day 24. Data were expressed as mean ± SEM of 8–10 mice/group (in two independent experiments). To perform quantitative reverse transcription-polymerase chain reaction (qRT-PCR) we have used 6 mice/group. To perform biochemical analysis in plasma we have used 8–10 mice/group. To perform ipGTT we have used 4 mice/group. To perform indirect calorimetry, we have used 4 mice/group. Two-way ANOVA followed by Sidak’s post hoc test and Mann–Whitney test were used for statistical analyses.
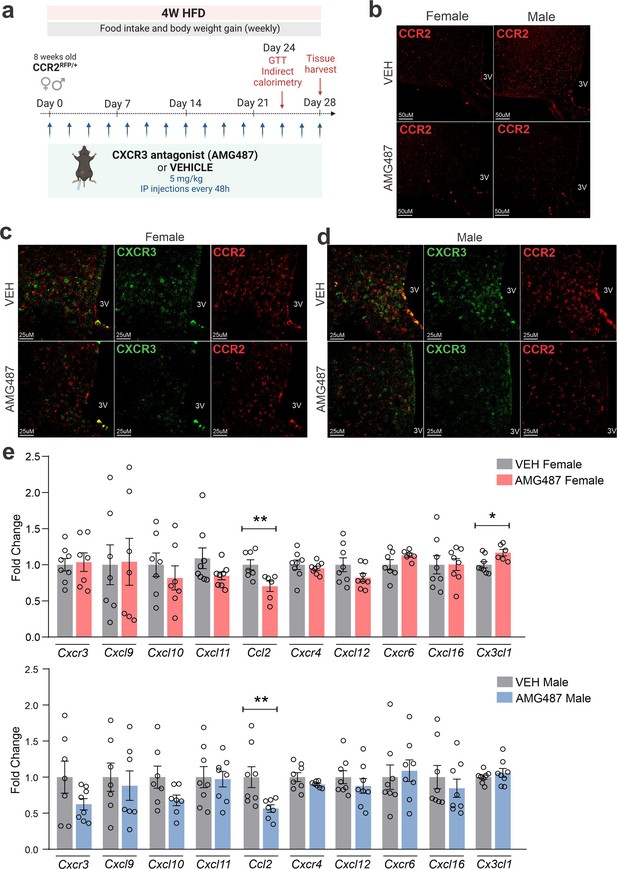
AMG487 treatment attenuates CCR2- and CXCR3-positive cell chemotaxis toward the hypothalamus of high-fat diet (HFD)-fed mice.
(a) Schematic representation of the experimental protocol for CXCR3 systemic blockage. (b) Coronal brain sections from 4 weeks HFD-fed CCR2RFP/+ mice showing the CCR2-positive cells distribution in the hypothalamic parenchyma upon AMG487 treatment. 3V = third ventricle, scale bars = 50 μm. (c) Coronal brain sections from 4 weeks HFD-fed female CCR2RFP/+ mice immunostained for CXCR3 upon AMG487 treatment. (d) Coronal brain sections from 4 weeks HFD-fed male CCR2RFP/+ mice immunostained for CXCR3 upon AMG487 treatment. 3V = third ventricle, scale bars = 25 μm. (e) Hypothalamic mRNA levels of several chemokine receptors and chemokines in HFD-fed female (light gray and pink bars) and male (light gray and blue bars) CCR2RFP/+ mice upon AMG487 treatment. For qualitative confocal image analysis, we have used 3 samples per group. For RT-qPCR of the hypothalamus, we have used 7–8 samples per group. Two-tailed Mann–Whitney tests were used for statistical analyses. *p < 0.05 and **p < 0.01 in comparison with respective VEH-treated groups.
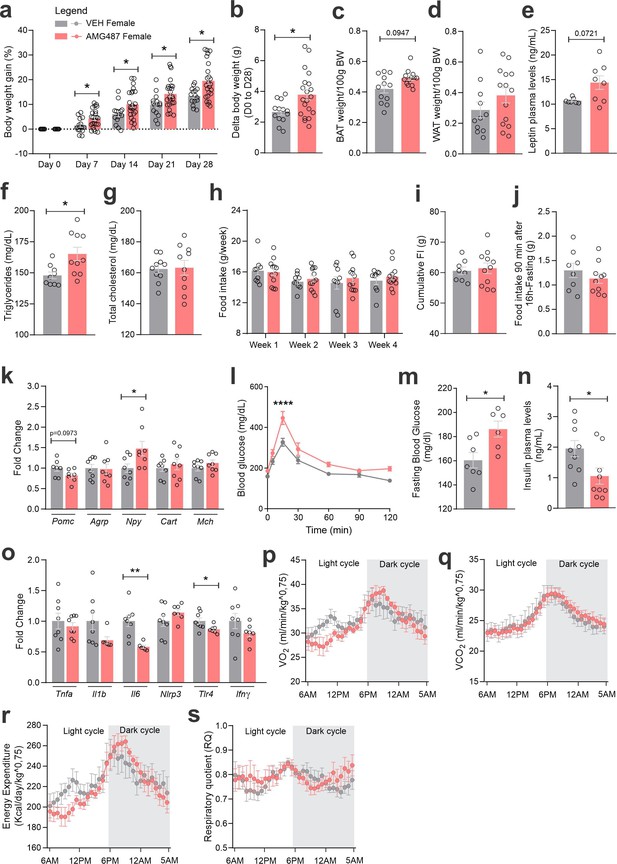
CXCR3 systemic blockage in high-fat diet (HFD)-fed female mice.
(a) Percentual of body weight gain from Day 0 to 28 of the experimental protocol. (b) Delta body weight during the experimental period. (c) Brown adipose tissue weight and (d) white adipose tissue (retroperitoneal depot) weight at Day 28. (e) Leptin, (f) triglycerides, and (g) total cholesterol plasma levels at Day 28. (h) Weekly food intake measurement during experimental period. (i) Cumulative food intake during the experimental period. (j) 90-min food intake measurement after 16-hr fasting. (k) Hypothalamic mRNA levels of neuropeptides involved in food intake control. (l) Intraperitoneal glucose tolerance test at Day 24. (m) 6-hr fasting blood glucose levels. (n) Insulin plasma levels at Day 28. (o) Hypothalamic mRNA levels of inflammatory genes. (p) O2 consumption; (q) CO2 production; (r) energy expenditure; (s) respiratory quotient at Day 24. Data were expressed as mean ± SEM of 14–16 mice/group (in four independent experiments). To perform quantitative reverse transcription-polymerase chain reaction (qRT-PCR) we have used 8 mice/group. To perform biochemical analysis in plasma we have used 8–10 mice/group. To perform ipGTT we have used 5 mice/group. To perform indirect calorimetry, we have used 4–5 mice/group. Two-way ANOVA followed by Sidak’s post hoc test and Mann–Whitney test were used for statistical analyses. *p < 0.05, **p < 0.01, ****p < 0.0001 in comparison with VEH-treated group.
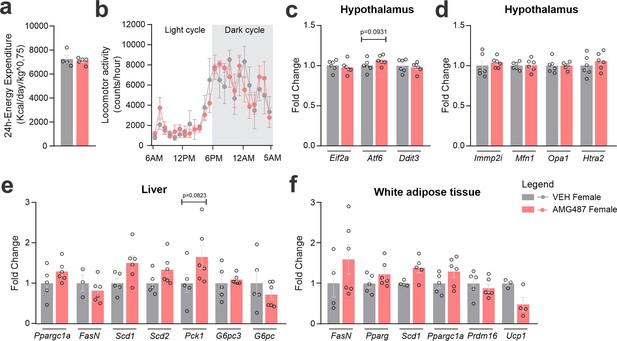
CXCR3 systemic blockage in high-fat diet (HFD)-fed female mice.
(a) Total 24-hr energy expenditure and (b) locomotor activity at Day 24 of the experimental protocol. (c) Delta body weight during the experimental period. (c) Hypothalamic mRNA levels of ER stress genes; (d) hypothalamic mRNA levels of mitochondrial function; (e) hepatic mRNA levels of genes of lipid and glucose metabolism; (f) white adipose tissue mRNA levels of genes of lipid metabolism and thermogenesis. Data were expressed as mean ± SEM of 4–6 mice/group. To perform indirect calorimetry and locomotor activity, we have used 5–6 mice/group. To perform quantitative reverse transcription-polymerase chain reaction (qRT-PCR) we have used 4–6 mice/group. Two-way ANOVA followed by Sidak’s post hoc test and Mann–Whitney test were used for statistical analyses.
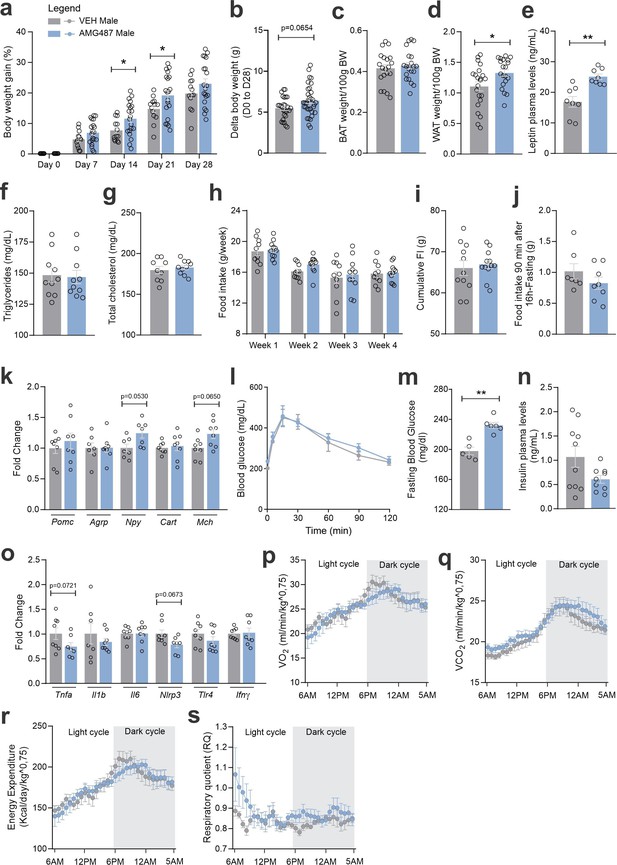
CXCR3 systemic blockage in high-fat diet (HFD)-fed male mice.
(a) Percentual of body weight gain from Day 0 to 28 of the experimental protocol. (b) Delta body weight during the experimental period. (c) Brown adipose tissue weight and (d) white adipose tissue (retroperitoneal depot) weight at Day 28. (e) Leptin, (f) triglycerides, and (g) total cholesterol plasma levels at Day 28. (h) Weekly food intake measurement during the experimental period. (i) Cumulative food intake during the experimental period. (j) 90-min food intake measurement after 16 hr of fasting. (k) Hypothalamic mRNA levels of neuropeptides involved in food intake control. (l) Intraperitoneal glucose tolerance test on Day 24. (m) 6-hr fasting blood glucose levels. (n) Insulin plasma levels at Day 28. (o) Hypothalamic mRNA levels of inflammatory genes. (p) O2 consumption; (q) CO2 production; (r) energy expenditure; (s) respiratory quotient at Day 24. Data were expressed as mean ± SEM of 14–16 mice/group (in four independent experiments). To perform quantitative reverse transcription-polymerase chain reaction (qRT-PCR) we have used 8 mice/group. To perform biochemical analysis in plasma we have used 8–10 mice/group. To perform ipGTT we have used 5 mice/group. To perform indirect calorimetry, we have used 4–5 mice/group. Two-way ANOVA followed by Sidak’s post hoc test and Mann–Whitney test were used for statistical analyses. *p < 0.05, **p < 0.01 in comparison with VEH-treated group.
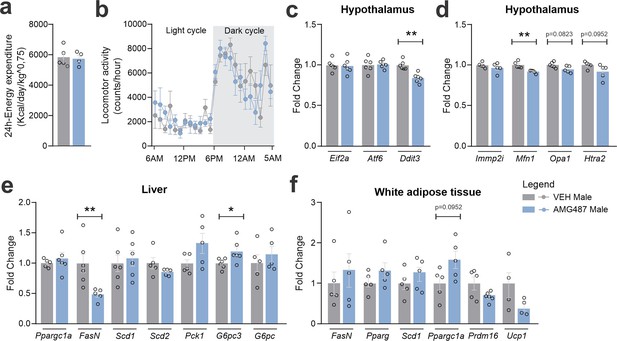
CXCR3 systemic blockage in high-fat diet (HFD)-fed male mice.
(a) Total 24-hr energy expenditure and (b) locomotor activity at Day 24 of the experimental protocol. (c) Delta body weight during the experimental period. (c) Hypothalamic mRNA levels of ER stress genes; (d) hypothalamic mRNA levels of mitochondrial function; (e) hepatic mRNA levels of genes of lipid and glucose metabolism; (f) white adipose tissue mRNA levels of genes of lipid metabolism and thermogenesis. Data were expressed as mean ± SEM of 4–6 mice/group. To perform indirect calorimetry and locomotor activity, we have used 5–6 mice/group. To perform quantitative reverse transcription-polymerase chain reaction (qRT-PCR) we have used 4–6 mice/group. Two-way ANOVA followed by Sidak’s post hoc test and Mann–Whitney test were used for statistical analyses. *p < 0.05 and **p < 0.01, in comparison with VEH-treated group.
Tables
Number of differentially expressed genes (DEGs) for each comparison.
| ↑ Up | ↓ Down | ||
|---|---|---|---|
| Effect of the diet in brain resident microglia cells | hfd_cx_f Cx3cr1_hfd_female vs. Cx3cr1_chow_female | 25 | 9 |
| hfd_cx_m Cx3cr1_hfd_male vs. Cx3cr1_chow_male | 261 | 151 | |
| Effect of the sex in brain resident microglia cells | chow_cx Cx3cr1_chow_male vs. Cx3cr1_chow_female | 9 | 11 |
| hfd_cx Cx3cr1_hfd_male vs. Cx3cr1_hfd_female | 7 | 5 | |
| Brain resident microglia cells vs. recruited myeloid cells | hfd_cc_f Cx3cr1_hfd_female vs. Ccr2_hfd_female | 4036 | 3569 |
| hfd_cc_m Cx3cr1_hfd_male vs. Ccr2_hfd_male | 3838 | 3350 | |
| Effect of the sex in recruited myeloid cells | hfd_cc Ccr2_hfd_male vs. Ccr2_hfd_female | 1598 | 1676 |





