Adult neurogenesis through glial transdifferentiation in a CNS injury paradigm
Figures

Neuropil glia proliferation in normal conditions and upon crush injury in adults.
(A) Schematic representation of Drosophila adult central nervous system (CNS) and ventral nerve cord (VNC), indicating the MtN, where the injury is performed. (B) Representative confocal image showing dividing cells (EdU+) with astrocyte-like glia (ALG) identity (Repo+ and Pros+) and glial identity different from ALG (Repo+, Pros-). (C) Number of ALG or ensheathing glia (EG) cells in G2-M per MtN, in normal conditions and 24 hr after injury. Paired t-test, *p<0.05, **p<0.01. (C’–C’’) Representative images of ALG in G2-M showing their position within the injured area (C’) and a magnification showing the detail of GFP+RFP+ cells (C’’). (D) Diagram of how the twin-spot mosaic analysis with a repressible cell marker (MARCM) tool works. (D’) Representative confocal image of ALG clones generated with twin-spot MARCM. (E) Number of VNCs with/without clones in controls and injured VNCs in young (1-day-old) and mature (7-day-old) animals. Chi-square binomial test, **p<0.01, ****p<0.0001. (F–G) Representative confocal images of a two-colour ALG clone (F) and a single-colour ALG clone (G). (H) Percentage of ALG single-colour clones in injured controls and injured VNCs where apoptosis was inhibited in ALG. Unpaired t-test, **p<0.01. Genotypes: wild-type (B); AlrmGal4>UAS-Fly-FUCCI (C-C’); R56F03Gal4>UAS-Fly-FUCCI (C); HsFLP; UAS-GFP, UAS-RFPRNAi /UAS-RFP, UAS-GFPRNAi; tubGal80ts:alrmGal4; (D’–H) HsFLP; UAS-GFP, UAS-RFPRNAi /UAS-RFP, UAS-GFPRNAi; tubGal80ts:alrmGal4/UAS-p35 (H). Scale bars: 50 μm (B, C, E); 15 μm (C’’, F, G).
-
Figure 1—source data 1
Nuclear sizes and position of transdifferentiated glial cells.
- https://cdn.elifesciences.org/articles/96890/elife-96890-fig1-data1-v1.xlsx
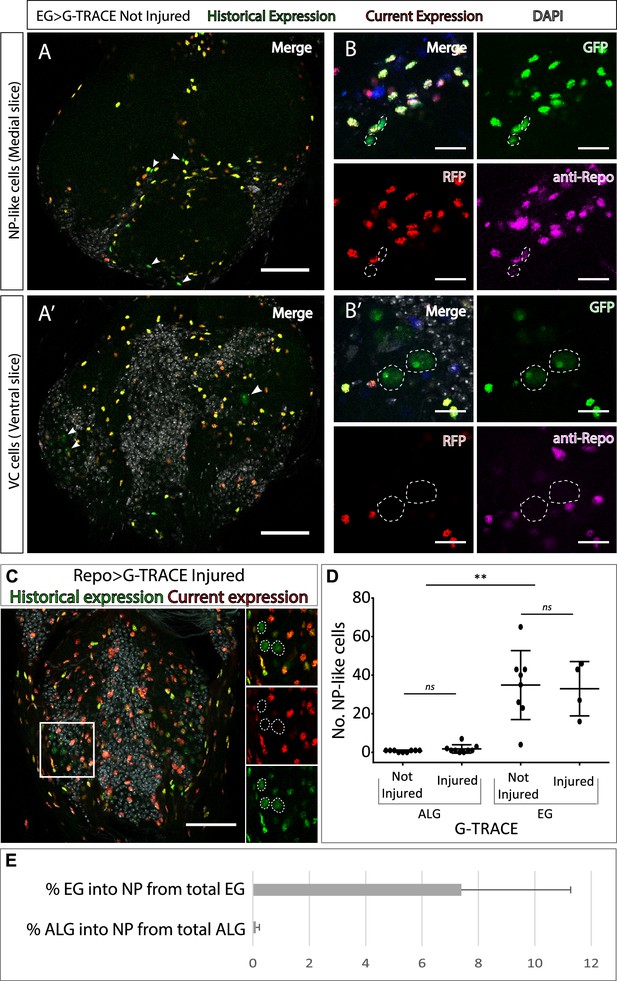
Neuropil glia transdifferentiate into two different cell types.
(A–B) Representative confocal slices showing transdifferentiated ensheathing glia (EG) cells with the Gal4 technique for real-time and clonal expression (G-TRACE) tool. (A) Medial slice showing neuropil (NP)-like cells position within the MtN. (B) Zoom slice (merge and separated channels) showing NP-like cells (GPF+, RFP-) have glial identity (repo+). (A’) Ventral slice showing ventral cortex (VC) cells position within the MtN. (B’) Zoom slice (merge and separated channels) showing VC cells (GPF+, RFP-) are not glia (repo-). (C) Representative ventral slice showing transdifferentiated CV cells visualised with G-TRACE toll driven by the pan-glial factor Repo-Gal4. (D) Number of NP-like cells originated from ALG or EG in not-injured and injured MtM. Mann-Whitney test ns p>0.05, **p<0.01. (E) Percentage of NP-like cells compared with the total number of NP glia that originate them. Scale bars: 50 μm (A, A’, C); 15 μm (B, B’). Genotypes: tubGal80ts, R56F03Gal4>UAS G-TRACE (A, B, D, E); tubGal80ts, repoGal4>UAS G-TRACE (C); tubGal80ts, AlrmGal4>UAS G-TRACE (D, E).
-
Figure 2—source data 1
Quantifications and statistic analysis for Figure 2.
- https://cdn.elifesciences.org/articles/96890/elife-96890-fig2-data1-v1.xlsx
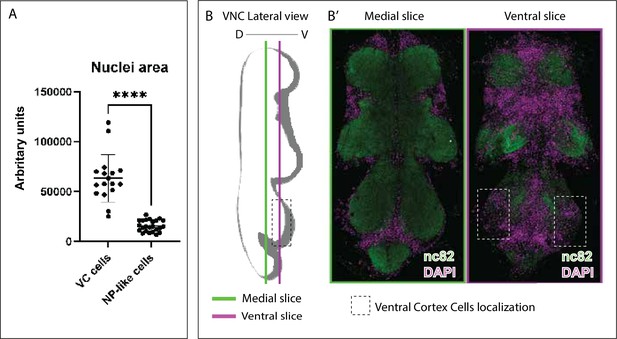
Nuclei sizes of transdifferentiated neuropil glia (NPG) and localisation of ventral cortex (VC) cells.
(A) Nuclei sizes of VC cells and neuropil (NP)-like cells. Paired t-test, ****p<0.0001. (B) Diagram showing VC cells localisation in a lateral ventral nerve cord (VNC) view. (B’) Frontal view of slices indicated in B. VNC of wild-type flies were stained with anti-nc82 (neuropil in green) and DAPI (nuclei/cortex in magenta). Genotype: Berlin (B’).
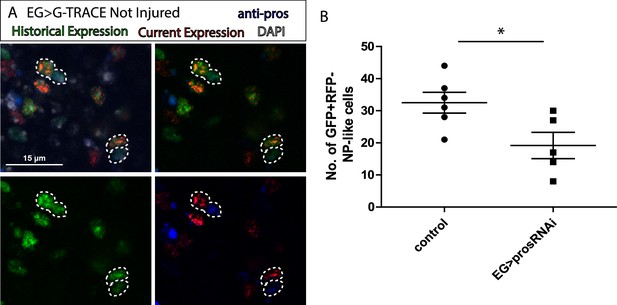
Ensheathing glia (EG) transdifferentiation into astrocyte-like glia (ALG) requires prospero.
(A) Representative image of neuropil (NP)-like cells originated from EG stained with ALG marker Prospero. Top left: Merge image with DAPI. Top right: Merge image without DAPI. Bottom left: GFP signal. Bottom right: RFP signal and pros staining. (B) Number of NP-like cells originated from EG in controls animals where pros was inhibited in EG. Unpaired t-test, *p<0.05. Scale bar: 15 μm (A). Genotypes: tubGal80ts, R56F03Gal4>UAS G-TRACE (A, B).
-
Figure 3—source data 1
Quantifications and statistic analysis for Figure 3.
- https://cdn.elifesciences.org/articles/96890/elife-96890-fig3-data1-v1.xlsx
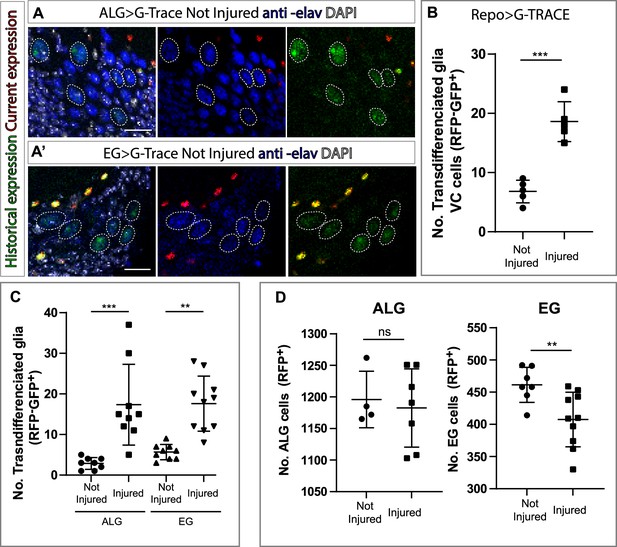
Ensheathing glia (EG) and astrocyte-like glia (ALG) transdifferentiate into neurons upon injury through two different mechanisms.
(A–A’) Transdifferentiated ventral cortex (VC) cells (GFP+, RFP-) raised from ALG (A) and EG (A’) are Elav+. From left to right: Merge with DAPI, merge without DAPI and green and red channels merged. (B) Number of transdifferentiated glia (VC cells, GFP+, RFP-) in controls and injured ventral nerve cords (VNCs). (C) Number of transdifferentiated VC cells (GFP+, RFP-) originates from ALG or EG in controls and injured mature animals. (D) Number of ALG or EG that remain ALG or EG in controls and injured VNCs. Statistics (B, C, D): Paired t-test, ***p<0.001, **p<0.01, ns p>0.05. Scale bar: 15 μm (A). Genotypes: tubGal80ts, R56F03Gal4>UAS G-TRACE and tubGal80ts, AlrmGal4>UAS G-TRACE.
-
Figure 4—source data 1
Quantifications and statistic analysis for Figure 4.
- https://cdn.elifesciences.org/articles/96890/elife-96890-fig4-data1-v1.xlsx

New neurons do not co-localise with markers for mature neurons.
Representative confocal images of ventral cortex (VC) cells (GFP+, RFP+) stained with DAPI (grey) and different antibodies in magenta, anti-ChAT (A), GluRIIA (B), and GABA (C). Dotted white circles indicate the VC cells. Scale bar: 15 μm (A). Genotypes: tubGal80ts, repoGal4>UAS G-TRACE.





