Nutritional state-dependent modulation of insulin-producing cells in Drosophila
Figures
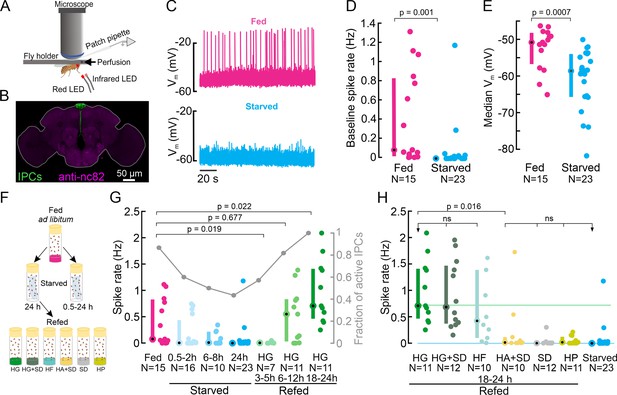
Insulin-producing cell (IPC) activity is modulated by the nutritional state and increases in response to feeding on nutritive sugars.
(A) Schematic of the setup for in vivo IPC whole-cell patch-clamp recordings. (B) IPCs in the Drosophila brain. UAS-myr-GFP was expressed under a Dilp2-GAL4 driver to label all 14 IPCs. The GFP signal was enhanced with anti-GFP (green), and brain neuropils were stained with anti-nc82 (magenta). (C) Representative examples of the membrane potential of two IPCs recorded in fed (magenta) and starved (cyan) flies. (D) Average baseline spike rate and (E) membrane potential of IPCs in fed (magenta) and starved (cyan) flies. (F) Schematic of the experimental starvation and refeeding protocol. HG: High glucose, HG + SD: High glucose with a standard diet, HF: High fructose, HA + SD: High arabinose with a standard diet, SD: Standard diet, HP: High protein. (G) Comparison of IPC spike rates in fed flies (magenta), increasingly starved flies (cyan), and flies refed on HG for different durations (green). Right axis shows a fraction of active IPCs (number of IPCs with spike rate >0 Hz, gray lines, and circles). (H) Comparison of IPC spike rate in flies refed on different diets. Reference lines represent median IPC activity in flies refed with HG for 18–24 h (green), and in flies starved for 24 h (cyan). Each circle represents an individual IPC, N=number of IPCs (see Supplementary file 1e for a number of flies), error bars indicate the median (circle) and inter-quartile range (IQR, bars). For D, E, and G, p-values are reported from the Wilcoxon rank-sum test. For H, a Kruskal-Wallis test followed by post hoc Wilcoxon rank-sum tests were used for pairwise comparisons. p-values are reported after the Holm-Bonferroni correction.
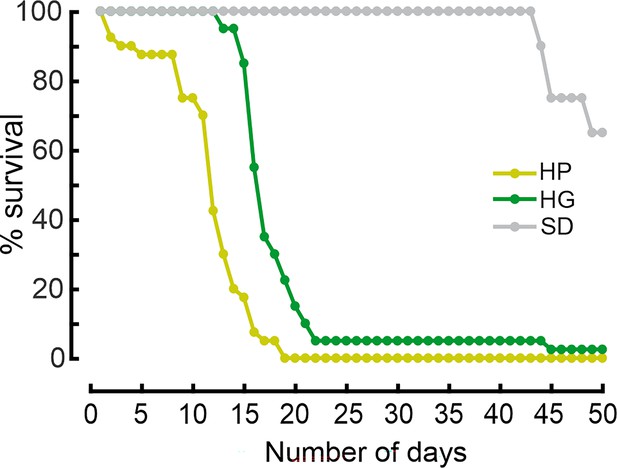
Dietary restriction impairs survival in Drosophila.
Percentage survival of flies on different diets. After 24 h of starvation, flies were kept on a high glucose (HG), high protein (HP), or standard diet (SD) and survival was scored every day. Data points were combined from two replicates of 20 flies per condition (N=40 flies per condition total).
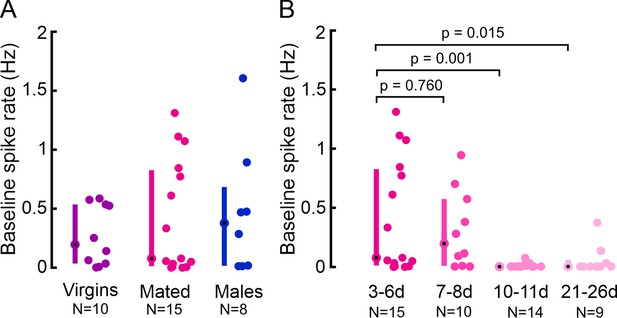
Sex and mating state do not affect insulin-producing cell (IPC) activity, but aging does.
(A) Comparison of IPC baseline spike rate in virgin females, mated females, and males. (B) Comparison of IPC baseline spike rate in different age groups. d=days, N=number of IPC recordings (see Supplementary file 1e for number of flies). Each dot represents one IPC, error bars indicate the median (circle) and interquartile range (bars). p-values calculated via the Wilcoxon rank-sum test.
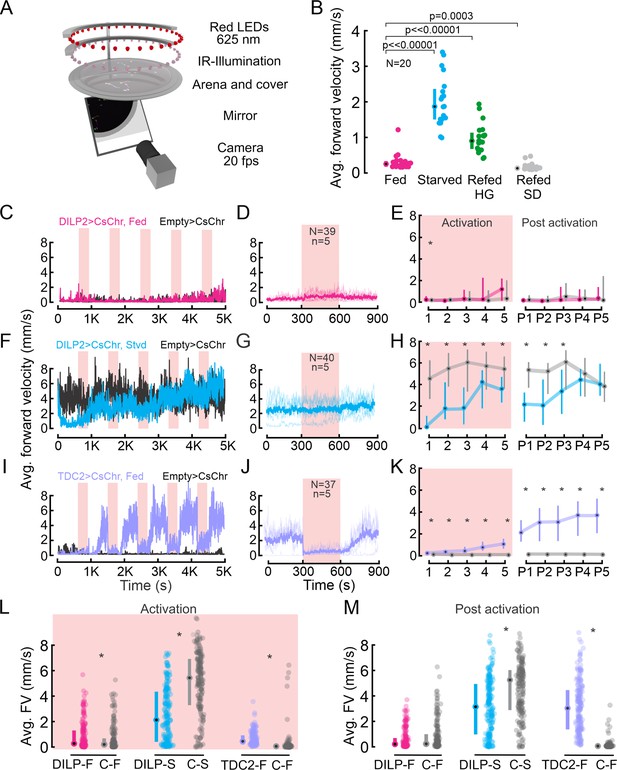
Walking activity is modulated by the nutritional state, insulin-producing cells (IPCs), and octopaminergic neurons (OANs).
(A) Schematic showing the Universal Fly Observatory (UFO) setup. (B) Average forward velocity (FV) of flies in different feeding states. Median and IQR are shown. p-values from the Wilcoxon rank-sum test. (C) Average FV of flies representing one replicate during optogenetic activation of IPCs in fed flies (magenta). Empty split-GAL4 was used as the control for all experiments (black). Red shading, optogenetic activation (300 s). (D) Average FV across all trials from two replicates for IPC activation in fed flies: 300 s before stimulus onset, during stimulus, and after stimulus offset. N=number of flies, n=number of activation trials. Thin lines represent individual trials, thick lines represent the median of all trials. (E) Average FV of all flies during each stimulus trial (1-5) and post-stimulus trial 300 s window immediately after activation ceased, (P1–P5) for IPC activation in fed flies. (F) Average FV of flies representing one replicate during optogenetic activation of IPCs in starved flies (cyan). (G) Average FV across all trials from two replicates for IPC activation in starved flies (plot details as in D). (H) Average FV during individual trials for IPC activation in staved flies (plot details as in E). (I) Average FV of flies representing one replicate during optogenetic activation of OANs in fed flies (lilac). (J) Average of FV across all trials from two replicates for OAN activation in fed flies (plot details as in D). (K) Average FV during individual trials for OAN activation in fed flies (plot details as in E). (L, M) Average FV pooled across all stimulus (1-5) and post-stimulus trials (P1–P5), respectively. Median and IQR are shown. p-values are reported from the Wilcoxon rank-sum test. Where no detailed p-value is stated, asterisks represent a significant difference. See also Supplementary file 1a and b.
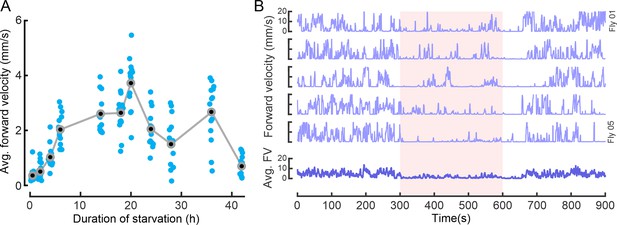
Starvation duration and octopaminergic neuron (OAN) activation affect locomotor activity.
(A) Average forward velocity of flies during different periods of starvation. N=20 flies per condition, except for 14 h (N=19), 20 h (N=19) and 28 h (N=16). Each point represents one fly, and gray circles and lines represent means. (B) Upper panel: Examples for forward velocities of five flies displaying stopping behavior during OAN activation (fifth activation cycle). Lower panel: Average forward velocity of all flies from one replicate (N=20). Red bar represents OAN activation.
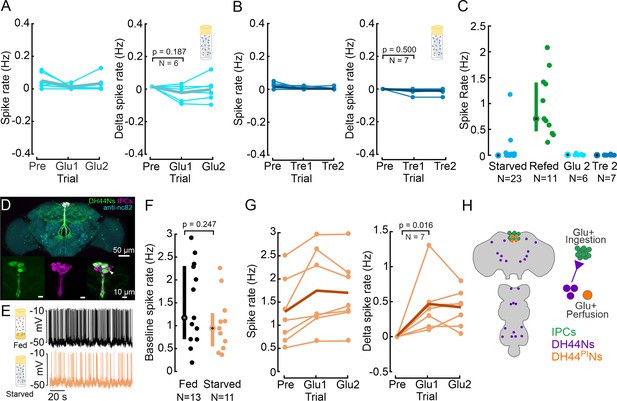
Insulin-producing cells (IPCs) are not sensitive to glucose perfusion but DH44PINs are.
(A) IPC spike rate and delta spike before and after perfusing extracellular saline with high glucose concentration, in starved flies. Baseline spike rates were recorded in glucose-free saline, followed by recordings in glucose-rich saline. Spike rates were averaged within a 5 min window. Delta spike rate was calculated by subtracting the baseline (Pre) from each trial for each IPC. Pre: 5 min recording in glucose-free saline. Glucose-rich saline was allowed to perfuse for about 8 min before analyzing IPC activity. Glu1 and Glu2: Two subsequent, 5 min long recordings in glucose-rich saline, starting eight minutes after onset of saline perfusion. Each circle represents an individual IPC from a different fly, the thick line represents the grand mean of all recordings. p-values are reported from the Wilcoxon signed-rank test. (B) Same experiment as in A, but with perfusion of trehalose-rich saline after recording the baseline. Note that our glucose-free saline was also devoid of trehalose (see methods). Other details same as A. (C) Comparison of IPC spike rate between starved, glucose-refed, glucose-perfused, and trehalose-perfused flies, highlighting the ‘incretin effect.’ Median and IQR are indicated. (D) Staining showing Drosophila brain with IPCs (magenta) and DH44Ns (green). UAS-myr-GFP was expressed under a DH44-GAL4 driver to label DH44 neurons. GFP was enhanced with anti-GFP (green), brain neuropils were stained with anti-nc82 (cyan), and IPCs were labeled using a Dilp2 antibody (magenta). White arrow indicates Dilp2 and DH44-GAL4 positive neurons. The other white regions in the image result from an overlap in z-projections between the two channels, rather than from antibody colocalization. (E) Example membrane potential of a DH44PIN recorded in a fed (black) and a starved fly (orange). (F) Baseline DH44PIN spike rate in fed (black) and starved flies (orange). Each circle represents an individual neuron (see Supplementary file 1f for a number of flies). Median and IQR are indicated. p-values are reported from Wilcoxon rank-sum test. (G) Spike rate and delta spike rate of DH44PINs before and during glucose perfusion in starved flies. Plot details same as A. (H) Schematic showing the location of cell bodies (left), and the regulation of IPCs, DH44PINs, and DH44Ns outside the pars intercerebralis (PI).
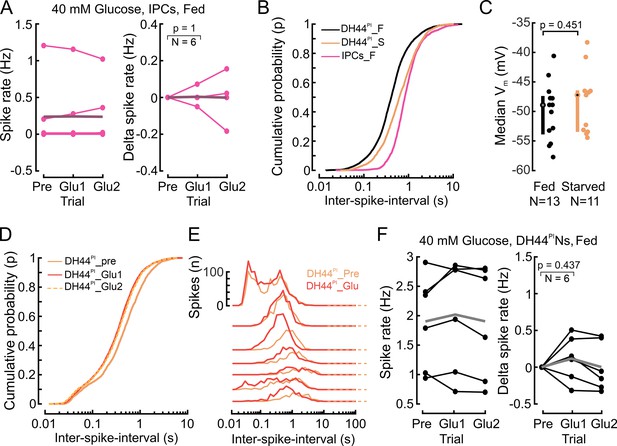
Glucose perfusion does not affect the activity of insulin-producing cells (IPCs) and DH44PINs in fed flies but shifts interspike interval distributions in DH44PINs.
(A) IPC spike rate (left) and change in spike rate (right) in fed flies under 40 mM glucose perfusion. Pre, 5 min recording in glucose-free extracellular saline, Glu1, and Glu2, two subsequent five-minute recordings in glucose-rich extracellular saline, starting eight minutes after the onset of glucose perfusion. Points represent the mean of individual flies, thick line represents the grand mean. p-values were calculated via the Wilcoxon signed-rank test. Delta spike rates were calculated by subtracting the mean spike rate during the Pre window. (B) DH44PIN spike activity patterns change with the nutritional state. Cumulative probability distribution of the ISI in DH44PINs and IPCs. F=fed, S=starved flies. Distributions were compared using a two-sample Kolgomorov-Smirnov test (DH44PI_F vs IPCs_F: p=1.5e–216; DH44PI_F vs DH44PI_S: p=2.5e–86; DH44PI_S vs IPCs_F: p=7.4e-51). The IPC dataset used in these panels corresponds to IPC baseline activity in fed flies in Figure 1D (N=15). DH44PI dataset corresponds to DH44PIN baseline activity in fed (N=13) and starved flies (N=11) in Figure 3F. (C) Median membrane potential of DH44PINs in fed and starved flies. (D) Cumulative probability distribution of the DH44 ISI in Pre, Glu1, and Glu2 windows reveals that DH44PIN activity became more bursty during glucose perfusion. Distributions were compared using a two-sample Kolgomorov-Smirnov test (DH44PI_pre vs DH44PI_Glu1: p=6.5e–30; DH44PI_pre vs DH44PI_Glu2: p=2.3e–21; DH44PI_Glu1 vs DH44PI_Glu2: p=0.05, N=7). (E) DH44PIN spike activity patterns change during glucose perfusion. ISIs are shown for DH44PINs before (Pre) and during (Glu1 and Glu2) 40 mM glucose perfusion. (F) DH44PIN spike rate (left) and change in spike rate (right) in fed flies under 40 mM glucose perfusion. Plot details as in A.
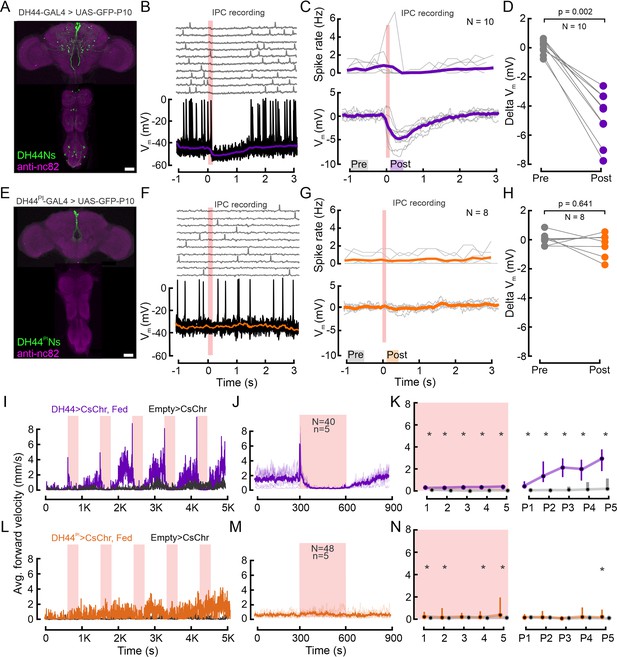
DH44Ns outside the pars intercerebralis (PI) inhibit insulin-producing cells (IPCs) and drive increases in locomotor activity.
(A) Immunolabeling showing DH44 expression in the brain and the VNC in the broad DH44-GAL4 driver line. GFP was enhanced with anti-GFP (green), and brain and VNC neuropils were stained with anti-nc82 (magenta). (B) Example recording of an IPC during optogenetic activation of the DH44Ns (red shading). Upper panel shows individual trials, lower panel shows ten trials overlaid, and the median of all trials (purple trace). (C) Upper panel shows the spike density of individual IPCs across 10 DH44N activations. Lower panel shows the baseline-subtracted, median-filtered Vm traces for each IPC. Thick lines represent the grand mean. (D) Effect of DH44N activation on IPCs. Delta Vm is plotted by calculating the median baseline subtracted Vm from C) 500 ms before (Pre) and 200 ms after DH44N activation (Post). Each circle represents one IPC recording. p-values from the Wilcoxon signed-rank test. (E) Immunolabeling showing GFP expression in the brain and the VNC in the sparse DH44PI-GAL4 driver line. (F) Example recording of an IPC during optogenetic activation of DH44 neurons using the sparse DH44PIN driver line. Plot details as in B. (G) Spike density and baseline subtracted median Vm of individual IPCs during activation of the DH44PIN driver line. Plot details as in C. (H) Pre and post-delta Vm of IPCs before and after optogenetic activation of the DH44PIN line. Plot details as in D. See Supplementary file 1e for a number of flies. (I) Average forward velocity (FV) of 20 flies during optogenetic activation of the DH44Ns using the broad driver line (see A). Empty split-GAL4 was used as a control for all experiments (black). Red shading indicates 300 s optogenetic activation windows. (J) Average FV across all DH44N activation trials based on two independent replications of the experiment in I. Note that the peak in average FV lies within the first frame of the stimulation window. (K) Average FV of all flies during each stimulus trial (1-5) and post-stimulus trial (300 s window immediately after activation ceased, (P1–P5). Circles and bars show median and IQR, respectively. Asterisks represent a significant difference according to a Wilcoxon rank-sum test. (L–N) Behavioral effects of optogenetic DH44PIN activation (see E). Plot details as in I, J, and K, respectively. See Supplementary file 1c and d for p-values. Scale bars: 50 µm.
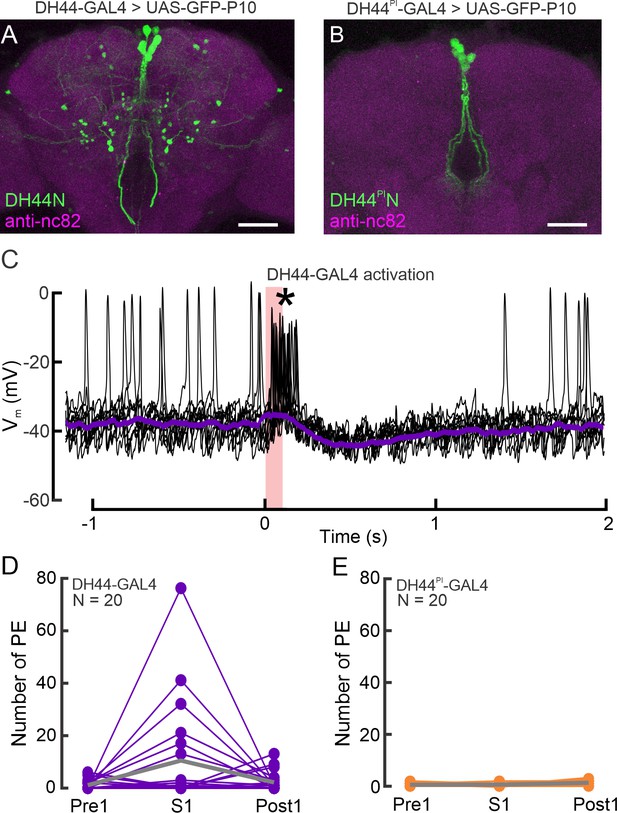
Effects of differential activation of DH44Ns and DH44PINs on insulin-producing cells (IPCs) and behavior.
(A) Expression pattern of the broad DH44N driver line in the brain visualized with a UAS-GFP-P10 reporter. GFP was enhanced with anti-GFP (green), and brain neuropils were stained with anti-nc82 (magenta). (B) Expression pattern of the sparse DH44PIN driver line. Staining details as in A. (C) Example recording of an IPC during optogenetic activation of the broad DH44N driver line. In this one example, we observed strong activation of the IPC during DH44N activation (asterisk). About 100 ms after activation, the IPC was inhibited similar to all other recordings (see Figure 4B). (D) Number of proboscis extensions (PE) before (Pre1), during (S1), and after the first LED pulse (Post1) activating DH44Ns in freely walking flies in the Universal Fly Observatory (UFO). PEs were scored manually. Gray lines represent the grand mean. (E) Activation of DH44PINs did not drive proboscis extension (PE). Plot details as in D. Scale bars: 50 µm.
Videos
Behavioral effects of optogenetic octopaminergic neuron (OAN) activation on Drosophila locomotor activity in the Universal Fly Observatory (UFO).
Example video of OAN-activated flies walking in the UFO during the fifth activation cycle. The white box indicates when the optogenetic stimulation LED is on. The video includes one-minute before activation (P4 in Figure 2K), 5 min after activation (5 in Figure 2K), and 1-minute after activation (P5 in Figure 2K). During OAN activation, the forward velocity decreased significantly, including several halting episodes.
Behavioral effects of the optogenetic activation protocol on the locomotor activity of Empty split-Gal4 control flies in the Universal Fly Observatory (UFO).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain (D. melanogaster) | DILP2-GAL4; CyO | Bloomington Drosophila Stock Center | BDSC:37516 | Used to drive expression in IPCs |
| Strain (D. melanogaster) | 10XUAS-IVS-myr::GFP | Bloomington Drosophila Stock Center | BDSC:32197 | Used to express GFP |
| Strain (D. melanogaster) | TDC2-GAL4 | Bloomington Drosophila Stock Center | BDSC:9313 | |
| Strain (D. melanogaster) | DH44-GAL4 | Bloomington Drosophila Stock Center | BDSC:39347 | |
| Strain (D. melanogaster) | DH44PI-GAL4 | Bloomington Drosophila Stock Center | BDSC:51987 | |
| Strain (D. melanogaster) | R96A08-LexA-p65-vk37::LexOp-dilp2::GFP; 20x-UAS-CsChrimson-attp2/TM6b | Liessem et al., 2023 | As described in the publication | Used for optogenetics experiments to drive CsChrimson in Gal4 driver lines while simultaneously expressing GFP in IPCs |
| Strain (D. melanogaster) | 10X-UAS-GFP-P10 | Bloomington Drosophila Stock Center | BDSC:32201 | |
| Strain (D. melanogaster) | 20X-UAS-CsChrimson | Bloomington Drosophila Stock Center | BDSC:55134 | |
| Strain (D. melanogaster) | Empty split-GAL4 | Bloomington Drosophila Stock Center | BDSC:86738 | |
| Chemical compound | all-trans-retinal | Sigma-Aldrich | R2500 | Used in optogenetic experiments |
| Software, algorithm | pCLAMP 10 | Molecular Devices | RRID:SCR_011323 | Used for electrophysiological recordings |
| Software, algorithm | MATLAB R2021a | The Mathworks | RRID:SCR_001622 | Used for data analysis and statistical testing |
| Software, algorithm | OCULAR | OCULAR | RRID:SCR_024467 | Image acquisition software |
| Antibody | anti-DILP2 (primary Rabbit polyclonal) | A. Veenstra, Bordeaux, FR | RRID:AB_2569969 | Used to label IPCs, diluted 1:2000 |
| Antibody | anti-nc82 (primary Mouse monoclonal) | DSHB | RRID:AB_2314866 | Used to label neuropils, diluted 1:500 |
| Antibody | anti-GFP (primary Chicken polyclonal) | Abcam | ab13970, RRID:AB_300798 | Enhances GFP signal, diluted 1:1000 |
| Chemical compound | Vectashield Antifade Mounting Medium | VEC-H-1000 | Biozol | |
| Antibody | Goat anti-chicken Alexa Fluor 488 (secondary goat polyclonal) | Thermo Fisher Scientific | RRID:AB_2534096 | 1:200 |
| Antibody | Goat anti-rabbit Alexa Fluor 555 (secondary goat polyclonal) | Thermo Fisher Scientific | RRID:AB_2535850 | 1:200 |
| Antibody | Goat anti-mouse Alexa Fluor 635 (secondary goat polyclonal) | Thermo Fisher Scientific | RRID:AB_2536184 | 1:400 |
| Chemical compound | SigmaCote | Sigma-Aldrich | cat. no. SL2; RRID:SCR_008988 | Siliconizing reagent |
| Software, algorithm | Fiji | Schindelin et al., 2012 | RRID:SCR_002285 | Used for image processing |
Additional files
-
Supplementary file 1
Tables listing the p-values for statistical tests performed for data sets shown in the main figures, along with the specific numbers of individual neurons and flies analyzed for each experiment.
- https://cdn.elifesciences.org/articles/98514/elife-98514-supp1-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/98514/elife-98514-mdarchecklist1-v1.pdf





