Seed Germination: Coping with salt stress
The growth of a seed into a seedling, a process known as germination, is the first stage of the plant life cycle. Seeds contain reserves of proteins and other nutrients that must be broken down into small molecules to provide raw materials and energy for this process. One of the most important raw materials is nitrogen, which is made available through a process called nitrogen remobilization (Tan-Wilson and Wilson, 2012). However, various environmental factors, such as temperature and soil conditions, can affect the timing of germination, and many questions about the relationship between nitrogen remobilization and germination remain unanswered.
It has been proposed that salt or a plant hormone called abscisic acid can inhibit germination by blocking the breakdown or degradation of the reserves of proteins and other nutrients that are stored within the seed (Kazachkova et al., 2016; Finkelstein and Lynch, 2000; Garciarrubio et al., 1997; Penfield et al., 2006). However, experiments have shown that certain reserves can be degraded in the presence of abscisic acid (Pritchard et al., 2002). Furthermore, evidence suggests that nitrogen remobilization during seed germination enhances protein degradation and the activities of enzymes like arginase and urease, particularly under salt stress. These enzymes have key roles in the conversion of the amino acid arginine into urea and then into ammonium, which acts as a source of nitrogen (Polacco et al., 2013; Siddappa and Marathe, 2020; Bu et al., 2015; Bu et al., 2024). Now, in eLife, Yuanyuan Bu (Northeast Forestry University), Shenkui Liu (Zhejiang A&F University) and colleagues report new insights into the mechanism by which salt can inhibit germination in Arabidopsis seeds (Bu et al., 2024; Figure 1).
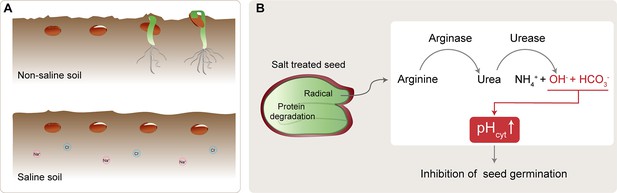
Salt-induced inhibition of seed germination.
(A) In non-saline soil conditions (top), seeds (brown circles) germinate and grow to become seedlings (green). In saline soil (bottom), which contains more sodium (Na+; pink) and chlorine (Cl-; blue) ions, seed germination is inhibited. (B) Under salt stress conditions, protein degradation in the seed creates arginine, which is converted to urea by arginase. The urea is subsequently hydrolyzed by urease, resulting in increased levels of ammonium ions (NH4+), bicarbonate ions (HCO3-), and hydroxide ions (OH-). The bicarbonate ions and hydroxide ions increase the cytoplasmic pH (pHcyt), thereby inhibiting seed germination.
First, the team inhibited enzymes required for the breakdown of arginine into urea using either chemical or genetic methods. This overcame the salt-induced inhibition of germination, suggesting excessive production of urea is a key contributor to the inhibition. To test whether accumulation of the ammonium from the subsequent urea breakdown was responsible for inhibiting germination, Bu et al. added ammonium to the medium the seeds were in. However, this did not have a negative impact on seed germination, challenging the long-held belief that ammonium accumulation and toxicity are the primary causes of salt-induced inhibition of seed germination (Bu et al., 2015).
Bu et al. next considered other factors from the urea breakdown pathway that could contribute. As well as yielding ammonium, the pathway also produces hydroxide ions, which are alkaline. Measuring the cytoplasmic pH of seed cells in different conditions revealed that they are more alkaline under salt stress. Further experiments confirmed that this is due to excessive hydrolysis producing alkaline hydroxide ions (Figure 1B). Taken together, the findings demonstrate that salt stress induces excessive hydrolysis of arginine-derived urea, resulting in an increase in the cytoplasmic pH of certain cells in the seed, which in turn prevents seed germination.
The findings of Bu et al. raise the question of the evolutionary benefit of plants retaining the urea hydrolysis pathway. A possible explanation is that this pathway acts as a survival mechanism. If seeds mature, fall to the ground, and encounter salty conditions, activating urea hydrolysis and inhibiting seed germination prevents seeds growing in unfavorable conditions. If the salt concentration then decreases, following rainfall for example, germination can proceed, ensuring the species survives. This adaptation may represent a form of ‘physiological dormancy’ in seeds.
Whether seed germination is influenced by external environmental factors or internal regulatory mechanisms, it fundamentally involves the regulation of the seed’s metabolism. It remains to be seen whether other environmental stresses – such as heat, drought, heavy metals, or plant hormones – also adhere to this principle. If it turns out to be universal, the implications for growing better crops could be profound. Understanding how seeds respond to various stresses would help us to both enhance crop resilience and develop sustainable agricultural practices, which will be crucial for increasing food security in the face of climate change.
References
-
Nickel and urease in plants: Still many knowledge gapsPlant Science 199–200:79–90.https://doi.org/10.1016/j.plantsci.2012.10.010
-
What we know about plant arginases?Plant Physiology and Biochemistry 156:600–610.https://doi.org/10.1016/j.plaphy.2020.10.002
-
Mobilization of seed protein reservesPhysiologia Plantarum 145:140–153.https://doi.org/10.1111/j.1399-3054.2011.01535.x
Article and author information
Author details
Publication history
Copyright
© 2024, Liang and Jiang
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Plant Biology
It is well documented that type-III effectors are required by Gram-negative pathogens to directly target different host cellular pathways to promote bacterial infection. However, in the context of legume–rhizobium symbiosis, the role of rhizobial effectors in regulating plant symbiotic pathways remains largely unexplored. Here, we show that NopT, a YopT-type cysteine protease of Sinorhizobium fredii NGR234 directly targets the plant’s symbiotic signaling pathway by associating with two Nod factor receptors (NFR1 and NFR5 of Lotus japonicus). NopT inhibits cell death triggered by co-expression of NFR1/NFR5 in Nicotiana benthamiana. Full-length NopT physically interacts with NFR1 and NFR5. NopT proteolytically cleaves NFR5 both in vitro and in vivo, but can be inactivated by NFR1 as a result of phosphorylation. NopT plays an essential role in mediating rhizobial infection in L. japonicus. Autocleaved NopT retains the ability to cleave NFR5 but no longer interacts with NFR1. Interestingly, genomes of certain Sinorhizobium species only harbor nopT genes encoding truncated proteins without the autocleavage site. These results reveal an intricate interplay between rhizobia and legumes, in which a rhizobial effector protease targets NFR5 to suppress symbiotic signaling. NFR1 appears to counteract this process by phosphorylating the effector. This discovery highlights the role of a bacterial effector in regulating a signaling pathway in plants and opens up the perspective of developing kinase-interacting proteases to fine-tune cellular signaling processes in general.
-
- Plant Biology
- Structural Biology and Molecular Biophysics
The Calvin-Benson-Bassham cycle (CBBC) performs carbon fixation in photosynthetic organisms. Among the eleven enzymes that participate in the pathway, sedoheptulose-1,7-bisphosphatase (SBPase) is expressed in photo-autotrophs and catalyzes the hydrolysis of sedoheptulose-1,7-bisphosphate (SBP) to sedoheptulose-7-phosphate (S7P). SBPase, along with nine other enzymes in the CBBC, contributes to the regeneration of ribulose-1,5-bisphosphate, the carbon-fixing co-substrate used by ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco). The metabolic role of SBPase is restricted to the CBBC, and a recent study revealed that the three-dimensional structure of SBPase from the moss Physcomitrium patens was found to be similar to that of fructose-1,6-bisphosphatase (FBPase), an enzyme involved in both CBBC and neoglucogenesis. In this study we report the first structure of an SBPase from a chlorophyte, the model unicellular green microalga Chlamydomonas reinhardtii. By combining experimental and computational structural analyses, we describe the topology, conformations, and quaternary structure of Chlamydomonas reinhardtii SBPase (CrSBPase). We identify active site residues and locate sites of redox- and phospho-post-translational modifications that contribute to enzymatic functions. Finally, we observe that CrSBPase adopts distinct oligomeric states that may dynamically contribute to the control of its activity.






