STAMBPL1 activates the GRHL3/HIF1A/VEGFA axis through interaction with FOXO1 to promote angiogenesis in triple-negative breast cancer
Figures
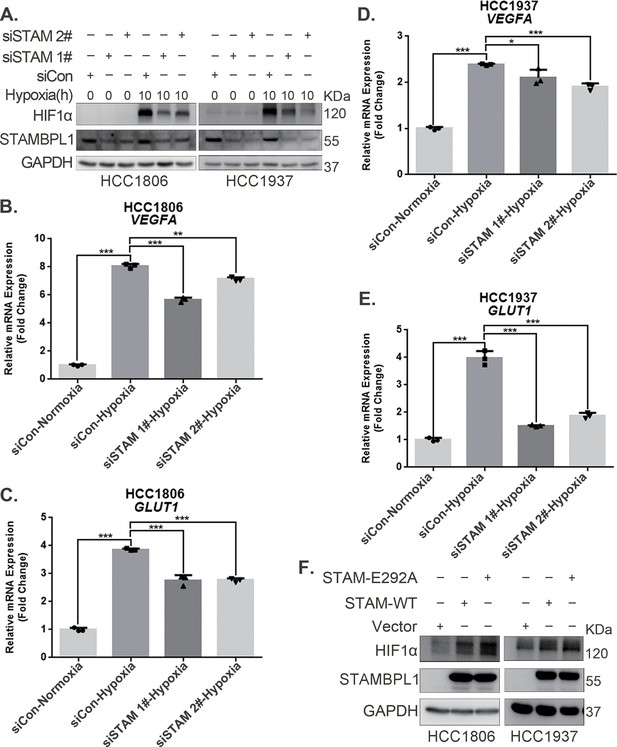
STAMBPL1 upregulates HIF1α in a non-enzymatic manner in triple-negative breast cancer (TNBC) cells.
(A) Western blot detected the protein levels of HIF1α when STAMBPL1 was knocked down under normoxia and hypoxia for 10 hr. (B, C) RNA samples were collected after 10 h of hypoxia treatment following the knockdown of STAMBPL1 in HCC1806 cells. RT-qPCR experiments were performed to detect the mRNA levels of the HIF1α downstream targets VEGFA and GLUT1 (n=3). (D, E) RNA samples of HCC1937 were collected as the above HCC1806 cells. RT-qPCR experiments were performed to detect the mRNA levels of the HIF1α downstream targets VEGFA and GLUT1 (n=3). (F) Western blot detected the protein levels of HIF1α when STAMBPL1/STAMBPL1-E292A was overexpressed under normoxia in HCC1806 and HCC1937 cells. Mean ± SD, *p < 0.05, **p < 0.01, ***p < 0.001, t-test.
-
Figure 1—source data 1
PDF file containing original western blots for Figure 1A, F, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/102433/elife-102433-fig1-data1-v1.zip
-
Figure 1—source data 2
Original files for western blot analysis displayed in Figure 1A, F.
- https://cdn.elifesciences.org/articles/102433/elife-102433-fig1-data2-v1.zip
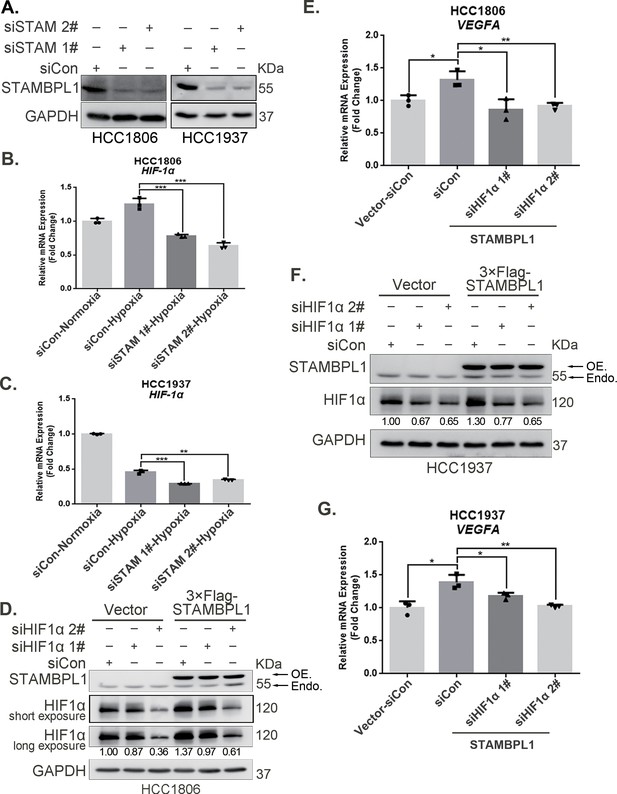
STAMBPL1 promotes HIF1A transcription and activates the HIF1α/VEGFA axis.
(A) Western blot analysis was performed to confirm the knockdown of STAMBPL1 in HCC1806 and HCC1937 cells. (B) RNA samples were collected after 10 hr of hypoxia treatment following the knockdown of STAMBPL1 in HCC1806 cells. RT-qPCR experiments were performed to detect the mRNA levels of HIF1α (n = 3). (C) RNA samples of HCC1937 cells were collected as the above HCC1806 cells. RT-qPCR experiments were performed to detect the mRNA levels of HIF1α (n = 3). (D) Knocking down HIF1α using siRNAs in HCC1806 cells stably overexpressing STAMBPL1 under normoxia. Western blot was used to detect the protein levels of STAMBPL1 and HIF1α. (E) RT-qPCR experiments were performed to detect the effect of HIF1α knockdown on the mRNA level of VEGFA mRNA in HCC1806 cells with STAMBPL1 overexpression under normoxia (n = 3). (F) Knocking down HIF1α using siRNAs in HCC1937 cells stably overexpressing STAMBPL1 under normoxia. Western blot was used to detect the protein levels of STAMBPL1 and HIF1α. (G) RT-qPCR experiments were performed to detect the effect of HIF1α knockdown on the mRNA level of VEGFA mRNA in HCC1937 cells with STAMBPL1 overexpression under normoxia (n = 3). Mean ± SD, *p < 0.05, **p < 0.01, ***p < 0.001, t-test.
-
Figure 2—source data 1
PDF file containing original western blots for Figure 2A, D, F, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/102433/elife-102433-fig2-data1-v1.zip
-
Figure 2—source data 2
Original files for western blot analysis displayed in Figure 2A, D, F.
- https://cdn.elifesciences.org/articles/102433/elife-102433-fig2-data2-v1.zip
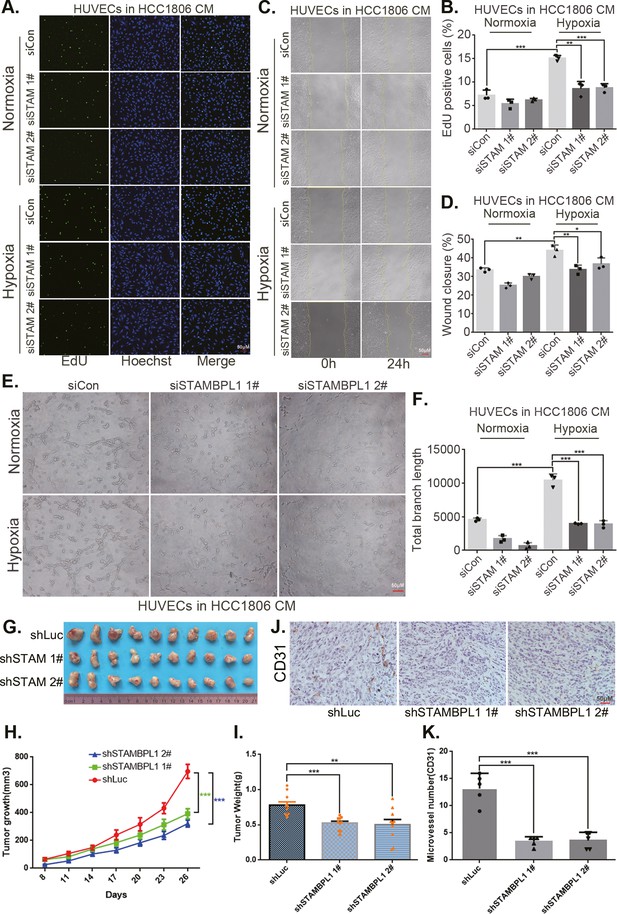
STAMBPL1 in triple-negative breast cancer (TNBC) cells enhances the activity of human umbilical vein endothelial cells (HUVECs) and promotes TNBC angiogenesis.
(A) EDU assay was performed to detect the effect of HCC1806 conditioned medium (CM) with STAMBPL1 knockdown under normoxia or hypoxia on the proliferation of HUVECs.The scale bar means 50 μm. (B) Statistical analysis of the EdU assay results (n = 3). The scale bar means 50 μm.(C) Wound healing assay was performed to detect the effect of HCC1806 CM with STAMBPL1 knockdown under normoxia or hypoxia on the migration of HUVECs. (D) Statistical analysis of the wound healing assay results (n = 3). (E) Tube formation assay was performed to detect the effect of HCC1806 CM with STAMBPL1 knockdown under normoxia or hypoxia on the tube formation of HUVECs. The scale bar means 50 μm. (F) Statistical analysis of the tube formation assay results (n = 3). (G) The growth of TNBC xenograft tumors was evaluated by photographing the tumors in nude mice to assess the role of STAMBPL1. (H, I) The growth and weight of the transplanted tumors in the nude mice were statistically analyzed, and the STAMBPL1sh#1 and STAMBPL1sh#2 groups were compared with the shLuc group (n = 10). (J) An immunohistochemical assay was used to detect the expression of the angiogenesis marker CD31 in xenograft tumors. The scale bar means 50 μm. (K) The number of microvessels in the immunohistochemical experiments was statistically analyzed (n = 5). Mean ± SD, *p < 0.05, **p < 0.01, ***p < 0.001, t-test.
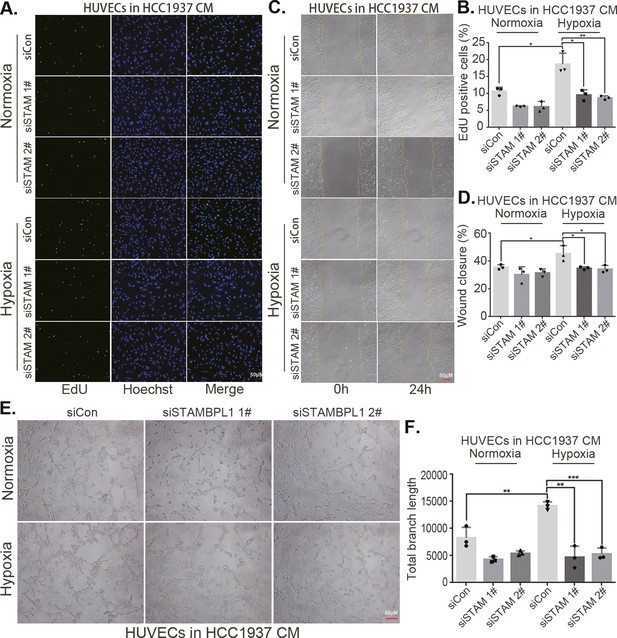
STAMBPL1 in triple-negative breast cancer (TNBC) cells enhances the activity of human umbilical vein endothelial cells (HUVECs).
(A) EDU assay was performed to detect the effect of HCC1937 conditioned medium (CM) with STAMBPL1 knockdown under normoxia or hypoxia on the proliferation of HUVECs. The scale bar means 50 μm. (B) Statistical analysis of the EdU assay results (n = 3). (C) Wound healing assay was performed to detect the effect of HCC1937 CM with STAMBPL1 knockdown under normoxia or hypoxia on the migration of HUVECs. The scale bar means 50 μm. (D) Statistical analysis of the wound healing assay results (n = 3). (E) Tube formation assay was performed to detect the effect of HCC1937 CM with STAMBPL1 knockdown under normoxia or hypoxia on the tube formation of HUVECs. The scale bar means 50 μm. (F) Statistical analysis of the tube formation assay results (n = 3). Mean ± SD, *p < 0.05, **p < 0.01, ***p < 0.001, t-test.
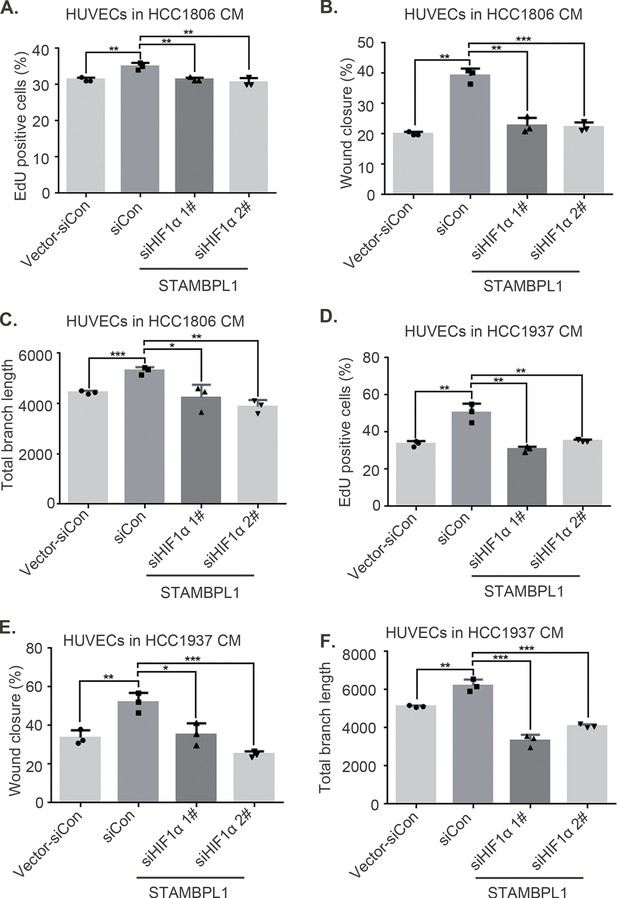
STAMBPL1 in triple-negative breast cancer (TNBC) cells enhances the activity of human umbilical vein endothelial cells (HUVECs).
(A, D) HCC1806 and HCC1937 cells were stably overexpressing STAMBPL1, and HIF1α was knocked down using siRNA. The conditioned medium (CM) was then collected and used to treat HUVECs. EdU assay was performed to detect the effect of CM with HIF1α knockdown on the proliferation of HUVECs as STAMBPL1 overexpression. Statistical analysis of the EdU assay results was performed. (B, E) Wound healing assay was performed to detect the effect of CM with HIF1α knockdown on the migration of HUVECs as STAMBPL1 overexpression. Statistical analysis of the wound healing assay results was performed. (C, F) Tube formation assay was performed to detect the effect of CM with HIF1α knockdown on the tube formation of HUVECs as STAMBPL1 overexpression. Statistical analysis of the tube formation assay results was performed. n = 3, Mean ± SD, *p < 0.05, **p < 0.01, ***p < 0.001, t-test.
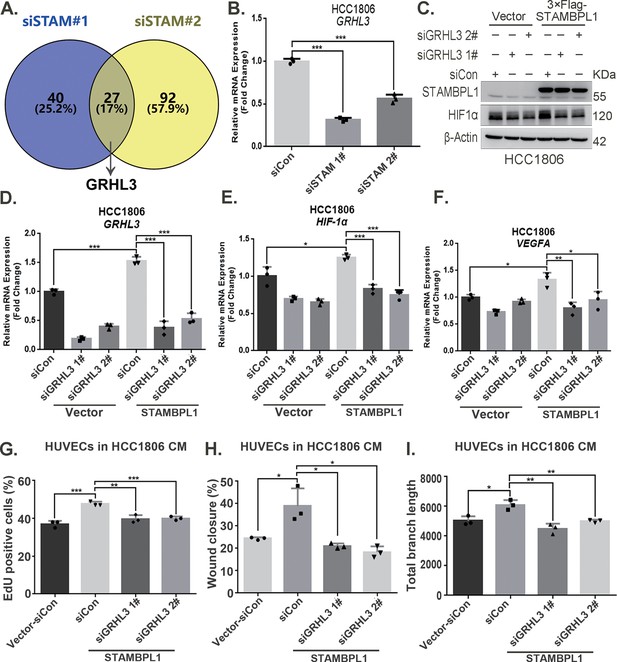
STAMBPL1 promotes HIF1A transcription via the upregulation of GRHL3.
(A) Venn diagram showing the overlap of differentially expressed genes identified via RNA-seq analysis. (B) RT-qPCR experiment was performed to detect the effect of STAMBPL1 knockdown on the mRNA levels of GRHL3 in HCC1806 cells (n = 3). (C) In HCC1806 cells stably overexpressing STAMBPL1, GRHL3 was knocked down via siRNA, after which the protein was collected. Western blotting was performed to detect the effect of GRHL3 knockdown on the expression of HIF1α as STAMBPL1 overexpression under normoxia. (D–F) RT-qPCR experiments were performed to detect the knockdown efficiency of GRHL3 and the effect of GRHL3 knockdown on the mRNA levels of HIF1α and its downstream target vascular endothelial growth factor A (VEGFA) when STAMBPL1 was overexpressing (n = 3). (G) Statistical analysis of the EdU assay results (n = 3). (H) Statistical analysis of the wound healing assay results (n = 3). (I) Statistical analysis of the tube formation assay results (n = 3). Mean ± SD, *p < 0.05, **p < 0.01, ***p < 0.001, t-test.
-
Figure 4—source data 1
PDF file containing original western blots for Figure 4C, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/102433/elife-102433-fig4-data1-v1.zip
-
Figure 4—source data 2
Original files for western blot analysis displayed in Figure 4C.
- https://cdn.elifesciences.org/articles/102433/elife-102433-fig4-data2-v1.zip
-
Figure 4—source data 3
Original files for RNA-seq analysis displayed in Figure 4A.
- https://cdn.elifesciences.org/articles/102433/elife-102433-fig4-data3-v1.zip
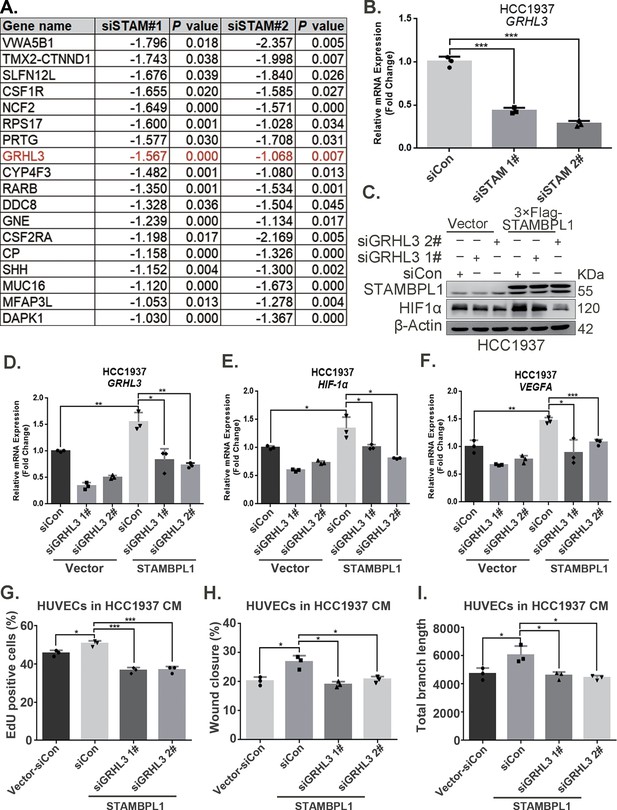
STAMBPL1 promotes HIF1A transcription via upregulating GRHL3.
(A) RNA-seq result: the list of 18 genes downregulated after STAMBPL1 knockdown in HCC1806 cells. (B) RT-qPCR experiments showed that knockdown of STAMBPL1 inhibited GRHL3 mRNA levels in HCC1937 cells. (C) In HCC1937 cells stably overexpressing STAMBPL1, GRHL3 was knocked down using siRNA, and then the protein was collected. Western blotting was performed to detect the effect of GRHL3 knockdown on the expression of HIF1α as STAMBPL1overexpression. (D–F) RT-qPCR experiments were performed to detect the knockdown efficiency of GRHL3 and the effect of GRHL3 knockdown on the mRNA levels of HIF1α and its downstream target vascular endothelial growth factor A (VEGFA) when STAMBPL1 was overexpressing. (G) Statistical analysis of EdU assay. (H) Statistical analysis of wound healing assay. (I) Statistical analysis of tube formation assay. n = 3, Mean ± SD, *p < 0.05, **p < 0.01, ***p < 0.001, t-test.
-
Figure 4—figure supplement 1—source data 1
PDF file containing original western blots for Figure 4—figure supplement 1C, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/102433/elife-102433-fig4-figsupp1-data1-v1.zip
-
Figure 4—figure supplement 1—source data 2
Original files for western blot analysis displayed in Figure 4—figure supplement 1C.
- https://cdn.elifesciences.org/articles/102433/elife-102433-fig4-figsupp1-data2-v1.zip
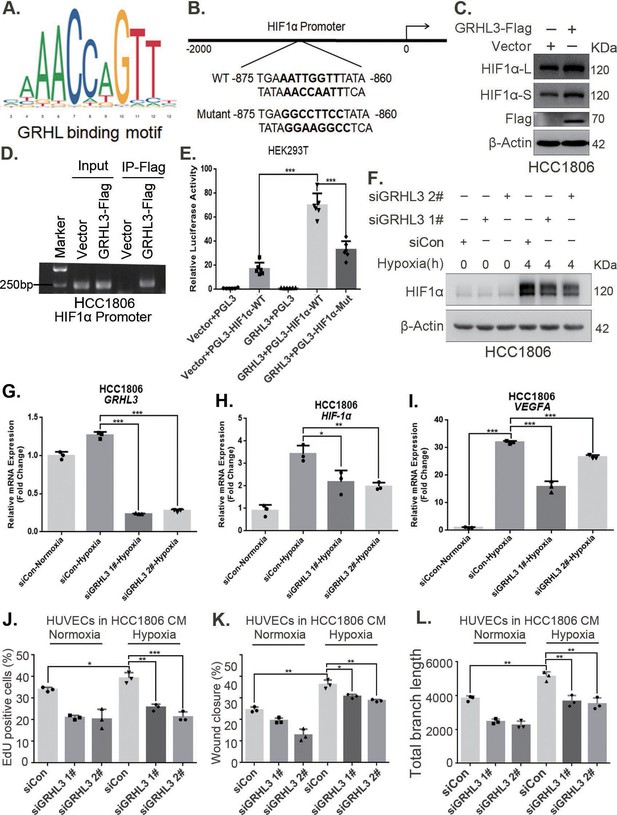
GRHL3 enhances HIF1A transcription by binding to its promoter.
(A) The JASPAR website was used to predict the potential binding sequence of the transcription factor GRHL3 to the HIF1α promoter. (B) Mutation pattern of the HIF1α promoter-binding sequence. (C) The protein level of HIF1α was detected in HCC1806 cells with GRHL3 overexpression by using western blot. (D) ChIP‒PCR experiments conducted in HCC1806 cells stably overexpressing GRHL3 to detect the interaction between GRHL3 and the promoter of HIF1A gene. (E) A luciferase assay was performed in HEK293T cells to detect the effect of GRHL3 on the transcriptional activity of HIF1A promoter (n = 6). (F) In HCC1806 cells, GRHL3 was knocked down by siRNA, and the cells were subjected to hypoxia for 4 hr. Western blotting was performed to detect the effect of GRHL3 knockdown on the protein level of HIF1α. (G–I) RT-qPCR experiments were used to detect the effect of GRHL3 knockdown on the mRNA levels of HIF1α and its downstream target vascular endothelial growth factor A (VEGFA) (n = 3). (J) In HCC1806 cells, GRHL3 was knocked down via siRNA, and the cells were then subjected to hypoxia for 24 hr. The conditioned medium (CM) was collected and used to treat human umbilical vein endothelial cells (HUVECs). EdU assays were performed to detect the effect of GRHL3 knockdown CM on the proliferation of HUVECs. Statistical analysis of the EdU assay results was performed (n = 3). (K) Wound healing assays were performed to detect the effect of GRHL3 knockdown CM on the migration of HUVECs. Statistical analysis of the wound healing assay results was performed (n = 3). (L) The tube formation assay was performed to detect the effect of GRHL3 knockdown CM on the tube formation of HUVECs. Statistical analysis of the tube formation assay results was performed (n = 3). Mean ± SD, *p < 0.05, **p < 0.01, ***p < 0.001, t-test.
-
Figure 5—source data 1
PDF file containing original western blots and DNA gels for Figure 5C, D, F, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/102433/elife-102433-fig5-data1-v1.zip
-
Figure 5—source data 2
Original files for western blot and DNA gel analysis displayed in Figure 5C, D, F.
- https://cdn.elifesciences.org/articles/102433/elife-102433-fig5-data2-v1.zip
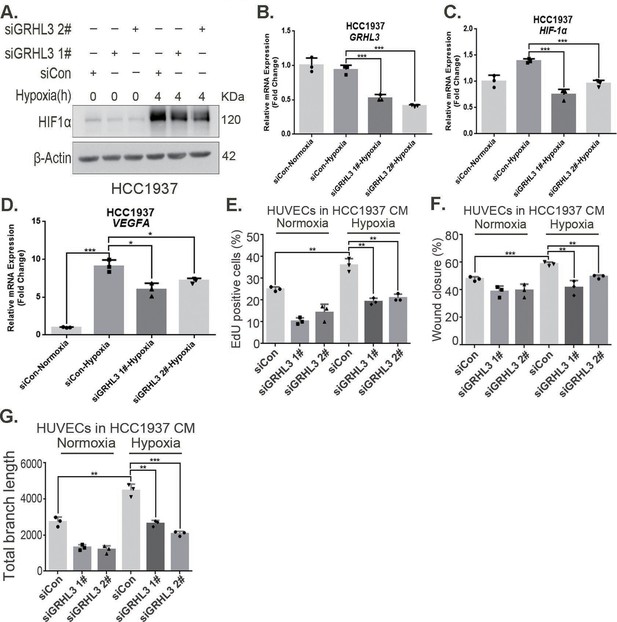
GRHL3 enhances HIF1A transcription by binding to its promoter.
(A) In HCC1937 cells, knockdown of GRHL3 using siRNA and treated with hypoxia for 4 hr. Western blotting was performed to detect the effect of GRHL3 knockdown on the protein level of HIF1α. (B–D) RT-qPCR experiments were performed to detect the knockdown efficiency of GRHL3 and the effect of GRHL3 knockdown on the mRNA levels of HIF1α and its downstream target vascular endothelial growth factor A (VEGFA). (E) In HCC1937 cells, GRHL3 was knocked down by siRNA and then treated with hypoxia for 24 hr, and then the conditioned medium (CM) was collected to treat human umbilical vein endothelial cells (HUVECs), EdU assay was performed to detect the effect of the CM with GRHL3 knockdown on the proliferation of HUVECs. (F) Wound healing assay was performed to detect the effect of the CM with GRHL3 knockdown on the migration of HUVECs. (G) The tube formation assay was performed to detect the effect of the CM with GRHL3 knockdown on the tube formation of HUVECs. n = 3, Mean ± SD, *p < 0.05, **p < 0.01, ***p < 0.001, t-test.
-
Figure 5—figure supplement 1—source data 1
PDF file containing original western blots for Figure 5—figure supplement 1A, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/102433/elife-102433-fig5-figsupp1-data1-v1.zip
-
Figure 5—figure supplement 1—source data 2
Original files for western blot analysis displayed in Figure 5—figure supplement 1A.
- https://cdn.elifesciences.org/articles/102433/elife-102433-fig5-figsupp1-data2-v1.zip
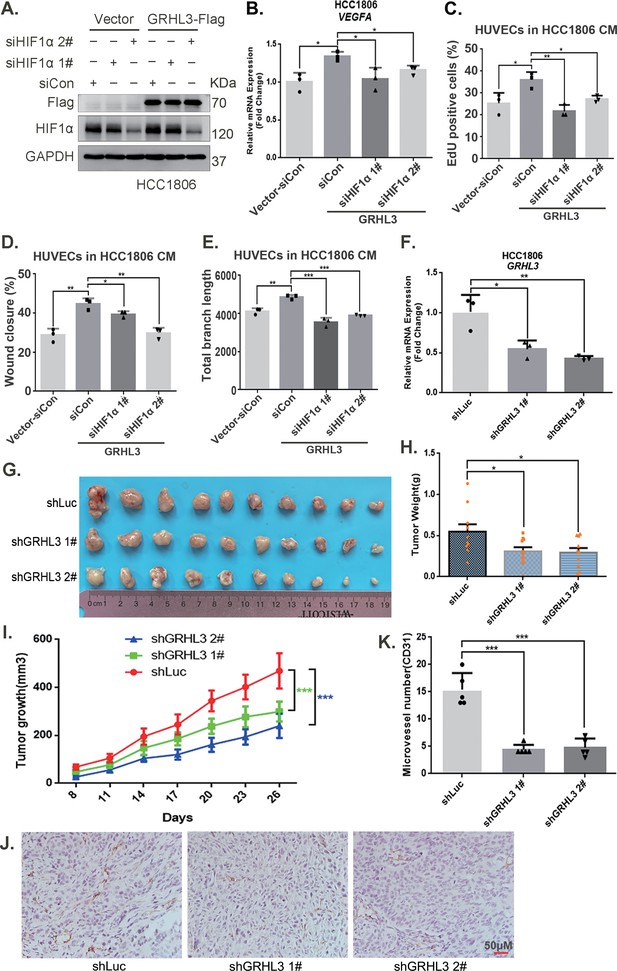
GRHL3 enhances HIF1A transcription by binding to its promoter.
(A) In HCC1806 cells stably overexpressing GRHL3, HIF1α was knocked down by siRNA, and the protein was subsequently collected. Western blotting was performed to detect the protein level of GRHL3-Flag and HIF1α. (B) RT-qPCR experiments were performed to detect the effect of HIF1α knockdown on the mRNA level of vascular endothelial growth factor A (VEGFA) as GRHL3 overexpression (n = 3). (C) HCC1806 cells stably overexpressing GRHL3 were used for the knockdown of HIF1α via siRNA. The conditioned medium (CM) was then collected and used to treat human umbilical vein endothelial cells (HUVECs). EdU assay was performed to detect the effect of CM with HIF1α knockdown on the proliferation of HUVECs as GRHL3 overexpression (n = 3). (D) Wound healing assay was performed to detect the effect of CM with HIF1α knockdown on the migration of HUVECs as GRHL3 overexpression (n = 3). (E) The tube formation assay was performed to detect the effect of CM with HIF1α knockdown on the tube formation of HUVECs as GRHL3 overexpression (n = 3). (F) RT-qPCR was used to detect the knockdown of GRHL3 in HCC1806 cells (n = 3). (G) Photographs of xenograft tumors from nude mice were taken to evaluate the role of GRHL3 in the growth of triple-negative breast cancer (TNBC) xenograft tumors. (H) The weights of the transplanted tumors from the nude mice were statistically analyzed, and the shGRHL3 1# and shGRHL3 2# groups were compared with the shLuc group (n = 10). (I) The growth of transplanted tumors in nude mice was statistically analyzed, and the shGRHL3 1# and shGRHL3 2# groups were compared with the shLuc group (n = 10). (J) An immunohistochemical assay was performed to detect the expression of the angiogenesis marker CD31 in xenograft tumors. The scale bar means 50 μm. (K) Statistical analysis of the number of microvessels in the immunohistochemical experiments (n = 5). Mean ± SD, *p < 0.05, **p < 0.01, ***p < 0.001, t-test.
-
Figure 6—source data 1
PDF file containing original western blots for Figure 6A, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/102433/elife-102433-fig6-data1-v1.zip
-
Figure 6—source data 2
Original files for western blot analysis displayed in Figure 6A.
- https://cdn.elifesciences.org/articles/102433/elife-102433-fig6-data2-v1.zip
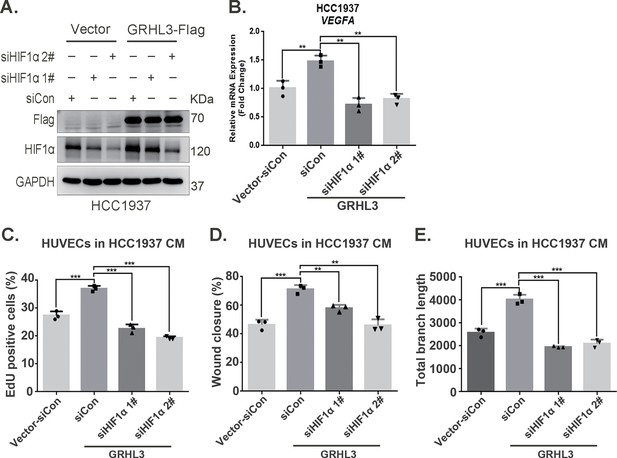
GRHL3 enhances HIF1A transcription by binding to its promoter.
(A) In HCC1937 cells stably overexpressing GRHL3, HIF1α was knocked down using siRNA, and then the protein was collected. Western blotting was performed to detect the protein levels of GRHL3 and HIF1α. (B) RT-qPCR experiment was performed to detect the effect of HIF1α knockdown on the mRNA expression of vascular endothelial growth factor A (VEGFA) as GRHL3 overexpression. (C) Statistical analysis of EdU assay results. (D) Statistical analysis of wound healing assay results. (E) Statistical analysis of tube formation assay results. n = 3, Mean ± SD, **p < 0.01, ***p < 0.001, t-test.
-
Figure 6—figure supplement 1—source data 1
PDF file containing original western blots for Figure 6—figure supplement 1A, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/102433/elife-102433-fig6-figsupp1-data1-v1.zip
-
Figure 6—figure supplement 1—source data 2
Original files for western blot analysis displayed in Figure 6—figure supplement 1A.
- https://cdn.elifesciences.org/articles/102433/elife-102433-fig6-figsupp1-data2-v1.zip
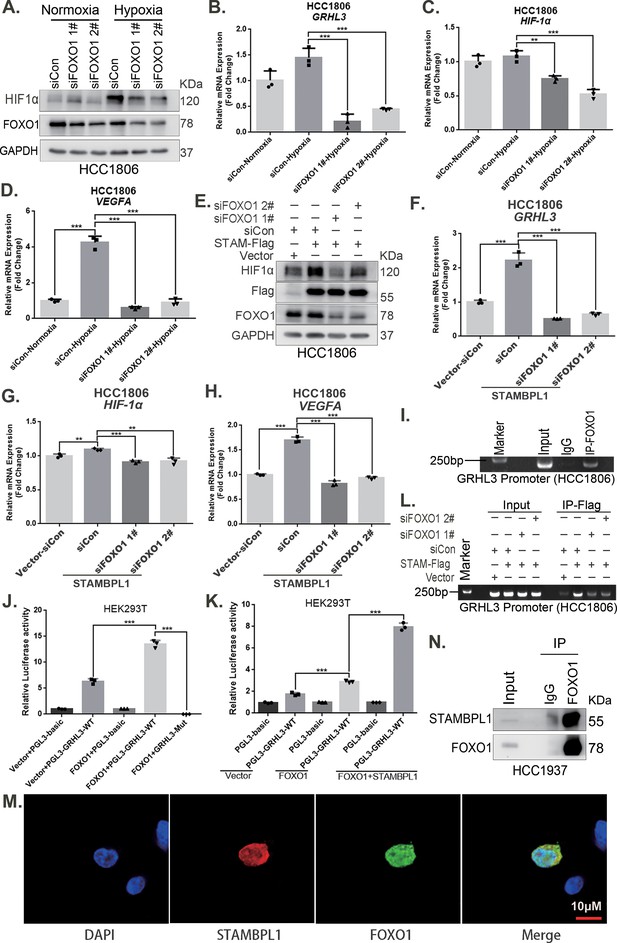
STAMBPL1 mediates GRHL3 transcription by interacting with FOXO1.
(A) Western blotting was used to detect the HIF1α protein expression when FOXO1 was knocking down in HCC1806 cells followed by hypoxia treatment for 4 hr. (B–D) RT-qPCR experiments were performed to detect the effect of FOXO1 knockdown on the mRNA levels of GRHL3/HIF1α/VEGFA in HCC1806 cells under hypoxia for 4 hr. (E) In HCC1806 cells stably overexpressing STAMBPL1, protein samples were collected after FOXO1 was knocked down via siRNA. Western blotting was performed to detect the effect of FOXO1 knockdown on the expression of HIF1α as STAMBPL1 overexpression. (F–H) RT-qPCR experiments were performed to detect the effect of FOXO1 knockdown on the mRNA expression of GRHL3, HIF1α and VEGFA as STAMBPL1 overexpression. (I) An endogenous ChIP‒PCR assay was performed using an anti-FOXO1 antibody in HCC1806 cells. (J) A luciferase assay was performed in HEK293T cells to detect the effect of FOXO1 on the transcriptional activity of GRHL3 promoter. (K) A luciferase assay was conducted in HEK293T cells to detected the effect of STAMBPL1 on the activation of the GRHL3 promoter by FOXO1. (L) ChIP‒PCR experiments were performed in HCC1806 cells stably overexpressing STAMBPL1 after knocking down FOXO1 via siRNA. (M) PCDH-STAMBPL1−3×Flag and PCDH-FOXO1−3×Flag plasmids were co-transfected into HEK293T cells, and then, immunofluorescence experiments were performed. Red represents STAMBPL1 staining, green represents FOXO1 staining, and blue represents DAPI staining. (N) Endogenous STAMBPL1 protein was immunoprecipitated from HCC1937 cells via an anti-FOXO1 antibody. Immunoglobulin G (IgG) served as the negative control. Endogenous STAMBPL1 was detected via western blotting.The scale bar means 10 μm. n = 3, Mean ± SD, **p < 0.01, ***p < 0.001, t-test.
-
Figure 7—source data 1
PDF file containing original western blots and DNA gels for Figure 7A, E, I, L, N, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/102433/elife-102433-fig7-data1-v1.zip
-
Figure 7—source data 2
Original files for western blot and DNA gel analysis displayed in Figure 7A, E, I, L, N.
- https://cdn.elifesciences.org/articles/102433/elife-102433-fig7-data2-v1.zip
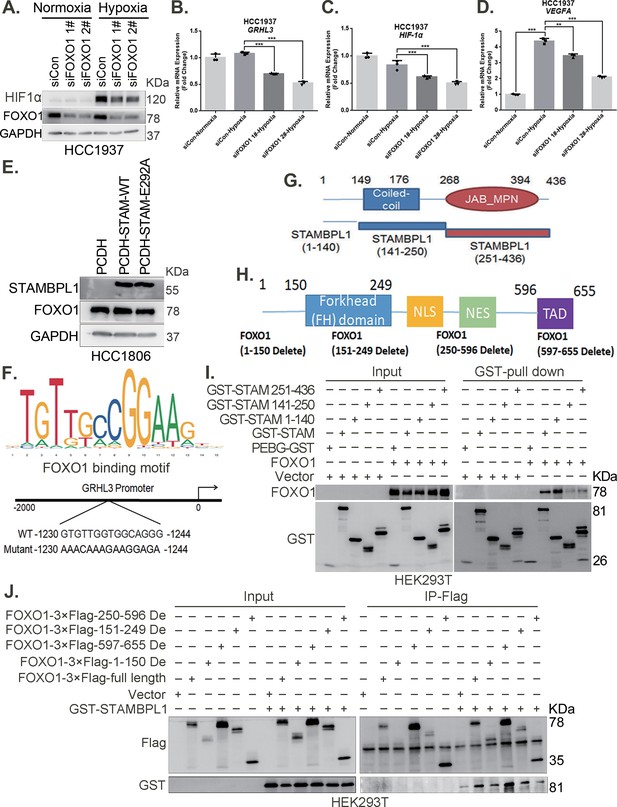
STAMBPL1 mediates GRHL3 transcription by interacting with FOXO1.
(A) Western blotting was performed to detect the effect of FOXO1 knockdown on the protein level of HIF1α in HCC1937 cells under hypoxia for 4 hr. (B–D) RT-qPCR experiments were performed to detect the effect of FOXO1 knockdown on the mRNA levels of GRHL3/HIF1α/VEGFA in HCC1937 cells under hypoxia treatment for 4 hr. (E) Western blot detected the FOXO1 expression when STAMBPL1 and STAMPBL1-E292A were overexpressed. (F) The JASPAR website predicts the possible binding sequence of transcription factor FOXO1 to the GRHL3 promoter and its mutation pattern map. (G) The truncated structure diagram of STAMBPL1. (H) The truncated structure diagram of FOXO1. (I) The full-length and truncated plasmids of pEBG-STAMBPL1-GST and pCDH-FOXO1-notag were transfected into HEK293T cells, and the proteins were collected for GST pull-down assay at 48 hr after transfection. (J) The full-length and truncated plasmids of pCDH-FOXO1-3×Flag and pEBG-STAMBPL1-GST were transfected into HEK293T cells, and the proteins were collected for IP-Flag assay at 48 hr after transfection. n = 3, Mean ± SD, **p < 0.01, ***p < 0.001, t-test.
-
Figure 7—figure supplement 1—source data 1
PDF file containing original western blots for Figure 7—figure supplement 1A, E, I, J, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/102433/elife-102433-fig7-figsupp1-data1-v1.zip
-
Figure 7—figure supplement 1—source data 2
Original files for western blot analysis displayed in Figure 7—figure supplement 1A, E, I, J.
- https://cdn.elifesciences.org/articles/102433/elife-102433-fig7-figsupp1-data2-v1.zip
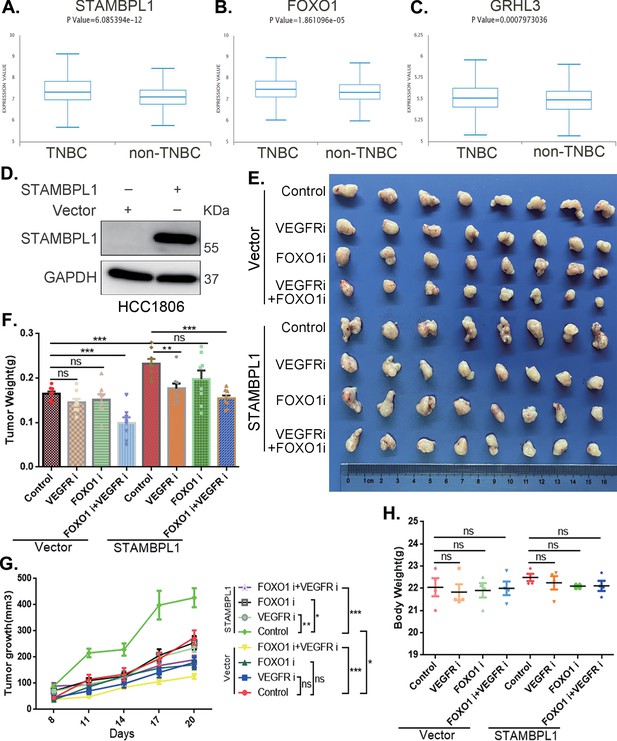
The combination of VEGFR and FOXO1 inhibitors synergistically suppresses triple-negative breast cancer (TNBC) xenograft growth.
(A–C) Metabric database analysis in BCIP revealed high expression levels of STAMBPL1, FOXO1, and GRHL3 in TNBC (n = 319) compared with non-TNBC (n = 1661). (D) The effect of STAMBPL1 overexpression in HCC1806 cells was detected by Western blotting. (E) Nude mice with tumors received the FOXO1 inhibitor AS1842856 (10 mg/kg, every 2 days) or the VEGFR inhibitor Apatinib (50 mg/kg, every 2 days) or a combination of both treatments. The effect of the drug treatments on the transplanted tumors was assessed by imaging the tumors. (F, G) The weight and growth of the transplanted tumors in the nude mice were statistically analyzed. The vector control group and the STAMBPL overexpression group were compared. The nondrug group and the combined drug group in the vector control group were compared. The nondrug group and the combined drug group in the STAMBPL overexpression group were compared (n = 8). (H) The final weights of the nude mice were statistically analyzed (n = 4). Mean ± SD, *p < 0.05, **p < 0.01, ***p < 0.001, t-test.
-
Figure 8—source data 1
PDF file containing original western blots for Figure 8D, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/102433/elife-102433-fig8-data1-v1.zip
-
Figure 8—source data 2
Original files for western blot analysis displayed in Figure 8D.
- https://cdn.elifesciences.org/articles/102433/elife-102433-fig8-data2-v1.zip
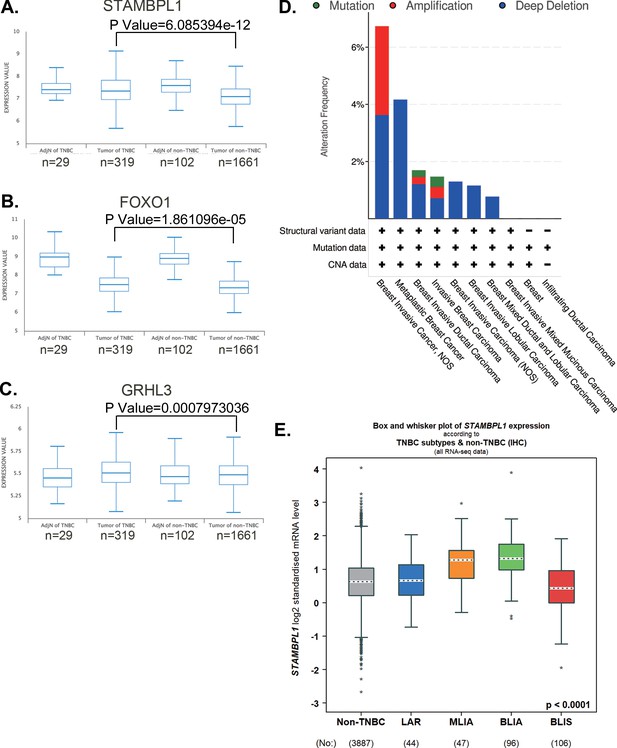
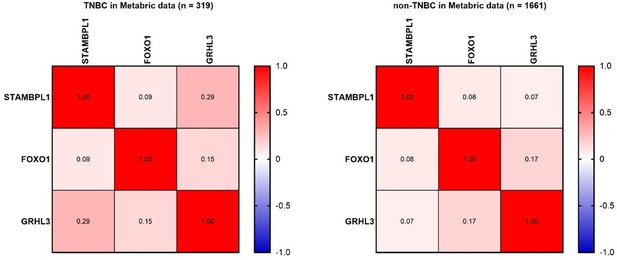
STAMBPL1, FOXO1, and GRHL3 is highly expressed in triple-negative breast cancer (TNBC).
(A–C) The expression of STAMBPL1, FOXO1, and GRHL3 in adjacent normal of TNBC (n = 29), TNBC (n = 319), adjacent normal of non-TNBC (n = 102), and non-TNBC (n = 1661) by analyzing the Metabric database in BCIP. (D) The mutation, amplification, and deep deletion of STAMBPL1 in breast cancer were analyzed by using cBioportal online database. (E) The expression of STAMBPL1 in non-TNBC and subtypes of TNBC by using bc-GenExMiner online database. Mean ± SD, *p < 0.0001, t-test.
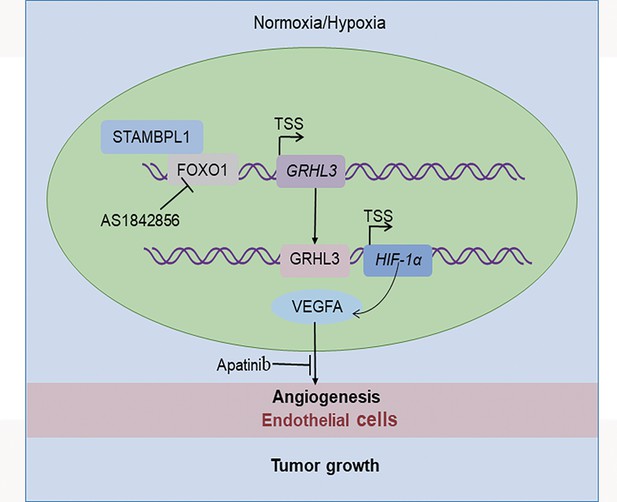
The working model of this study.
STAMBPL1 interacts with FOXO1 to promote triple-negative breast cancer (TNBC) angiogenesis by activating the GRHL3/HIF1α/VEGFA axis.