A systematic review and embryological perspective of pluripotent stem cell-derived autonomic postganglionic neuron differentiation for human disease modeling
Figures
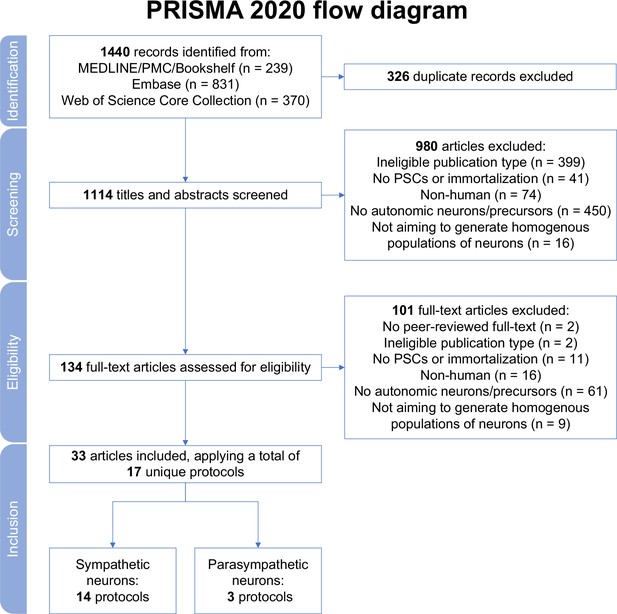
Study selection.
MEDLINE, Medical Literature Analysis and Retrieval System Online; PMC, PubMed Central; PRISMA, Preferred Reporting Items for Systematic reviews and Meta Analyses; PSCs, pluripotent stem cells.
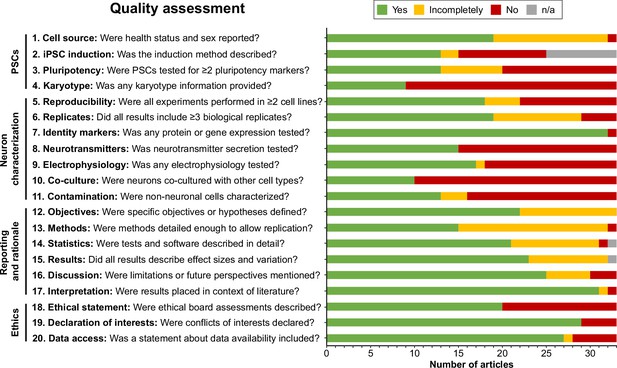
Quality assessment.
Quality assessment results per criterion. Criteria topics are indicated to the left of the criteria. See Figure 2—figure supplement 1 for results per article, and Supplementary file 2 for detailed criteria. iPSCs, Induced pluripotent stem cells; n/a, not applicable; PSCs, pluripotent stem cells.
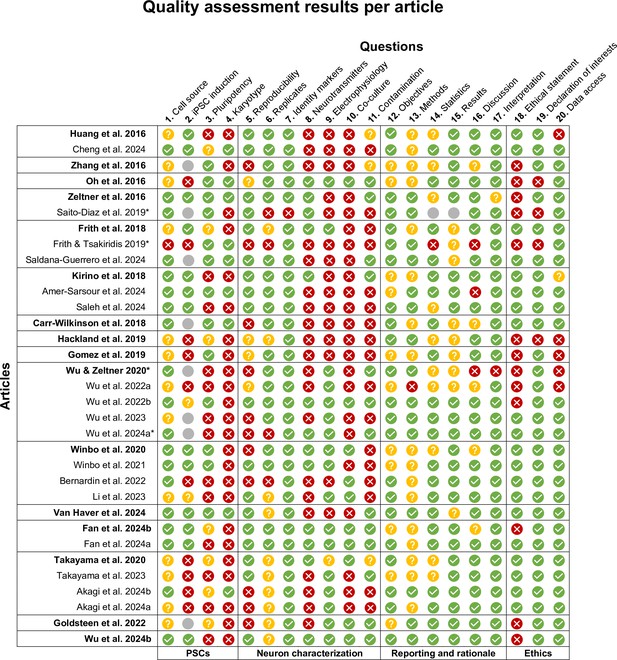
Quality assessment results per article.
Quality assessment results per article, grouped by protocol and neuron type, and listed chronologically. The earliest article per protocol is printed in bold. Reference numbers are indicated following the article authors and year. The quality criteria in Figure 2 have been indicated diagonally above each column. Topic categories have been indicated below the table. Please see Supplementary file 2 for detailed criteria. * Methodological articles were assessed using the same criteria as other articles. iPSCs, induced pluripotent stem cells; n/a, not applicable; PSCs, pluripotent stem cells.
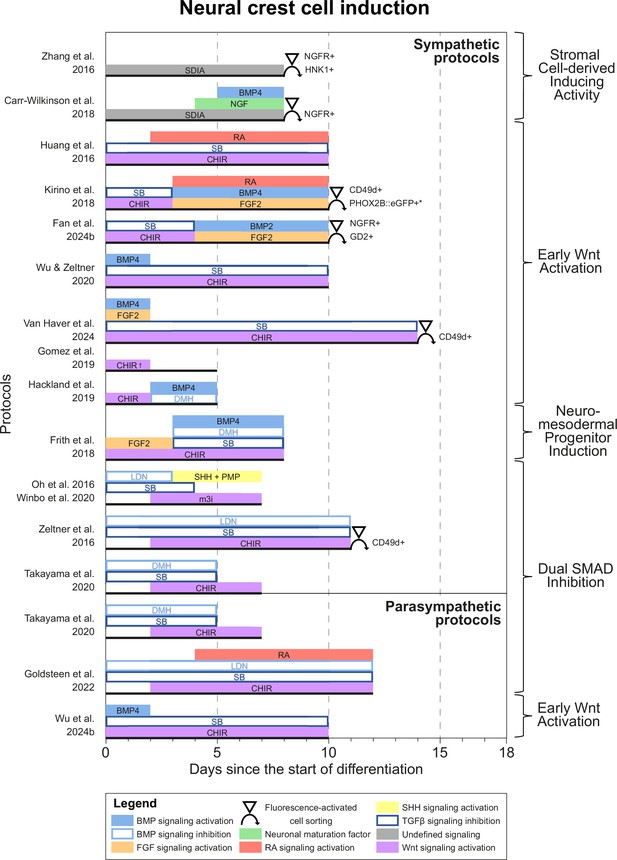
Neural crest cell induction.
Timings and signaling cues used during the first phase of differentiation until neural crest induction per unique protocol. Duration of this phase per protocol is indicated by the horizontal black bars. Categories of similar approaches are indicated to the right of the figure. Molecules targeting similar pathways have been grouped by color. Colors also match the signaling cues in Figures 4—6. * Selection step yields optimal cell purity, but this is not required. † Gomez et al., 2019 identified two optimal CHIR concentrations for neural crest induction, 3 µM and 10 µM. 10 µM was used for sympathetic neuron differentiation. BMP, bone morphogenetic protein; CD49d, Integrin subunit α4; CHIR, CHIR99021; DMH, dorsomorphin; eGFP, enhanced green fluorescent protein; FGF, fibroblast growth factor; GD2, disialoganglioside; HNK1, human natural killer-1; LDN, LDN193189; m3i, Modified three inhibitor approach (CHIR99021, DAPT, and PD173074); NGF, nerve growth factor; NGFR, nerve growth factor receptor; PHOX2B, paired-like homeobox 2b; PMP, purmorphamine; RA, retinoic acid; SB, SB431542; SDIA, stromal cell-derived inducing activity; SHH, Sonic hedgehog; TGFβ, transforming growth factor beta.
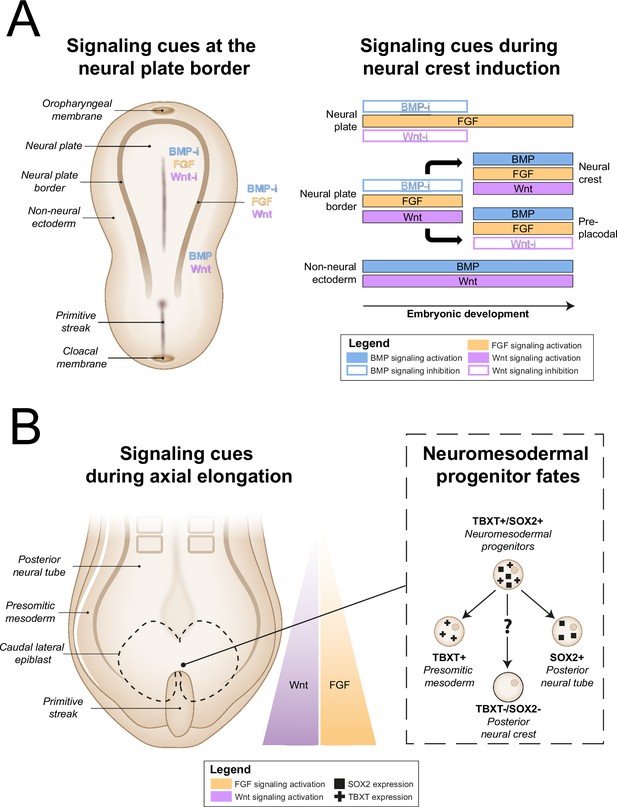
In vivo neural crest induction signaling requirements.
(A) General signaling requirements for distinct populations at the neural plate border. Left, a dorsal schematic view of the embryo and signaling cues (indicated in colored text) present near the neural plate border during early gastrulation are shown. Right, the temporal sequence of signaling cues required for distinct populations near the neural plate border in amniotes between gastrulation and neurulation is shown (based on Thawani and Groves, 2020). (B) Neuromesodermal progenitors arise in the tailbud during axial elongation under conditions of high Wnt and FGF signaling activation. Wnt and FGF concentrations form a rostrocaudal gradient, with highest concentrations in the tailbud. Right, neuromesodermal progenitors possibly contribute to posterior neural crest cell populations. BMP, bone morphogenetic protein; BMP-i, BMP signaling inhibition; FGF, fibroblast growth factor; SOX2, SRY-box transcription factor 2; TBXT, T-box transcription factor T; Wnt-i, Wnt signaling inhibition.
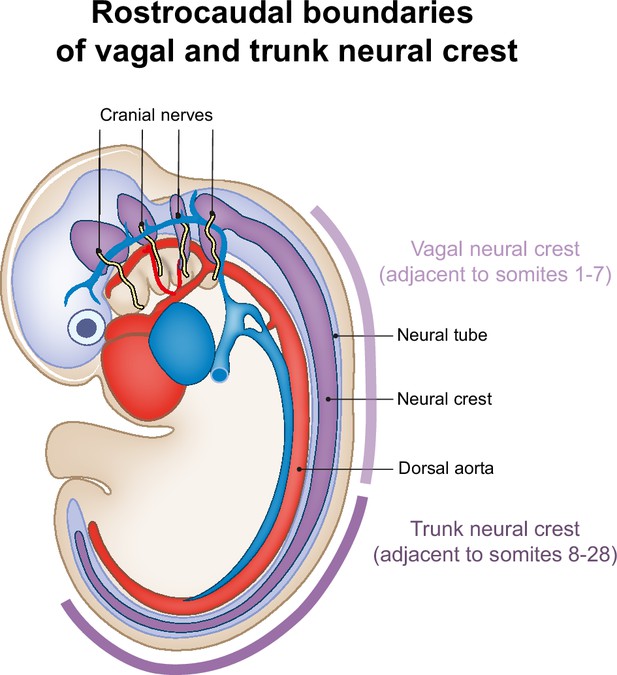
Rostrocaudal boundaries of vagal and trunk neural crest.
Lateral view of an embryo around the time of neural crest delamination. The rostrocaudal boundaries of vagal neural crest, which gives rise to parasympathetic neurons (Figure 6), are indicated by the light purple line adjacent to the embryo. The darker purple line indicates the rostrocaudal boundaries of trunk neural crest, which gives rise to sympathetic neurons (Figure 5). In later developmental stages, as axial elongation progresses, (sacral) neural crest continues to be formed caudally from trunk neural crest.
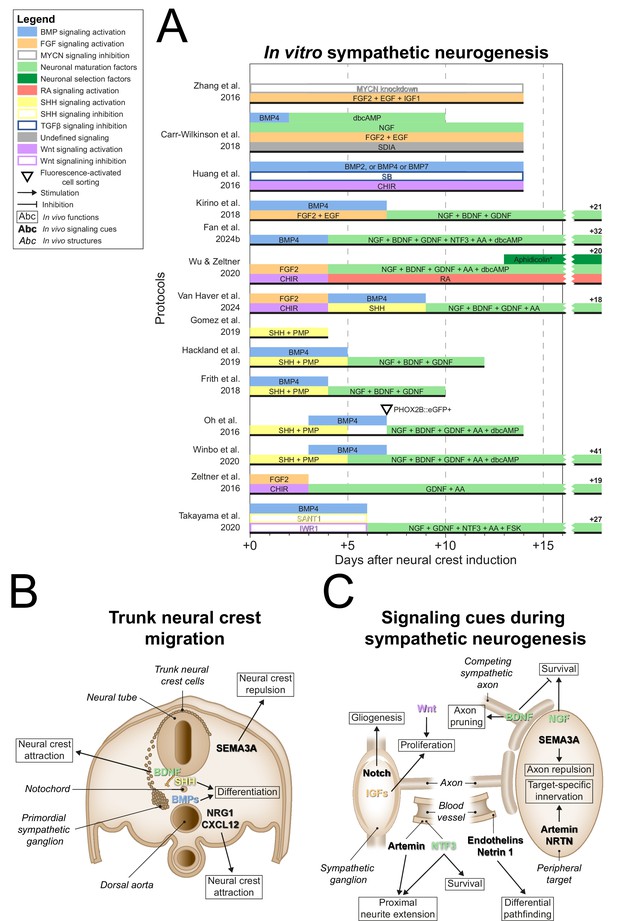
Sympathetic neurogenesis.
(A) Timings and signaling cues used from neural crest induction until the end of sympathetic neuron differentiation. Duration of this phase per protocol is indicated by the horizontal black bars. Total durations of this phase exceeding the width of the graph are indicated to the right of the graph. Molecules targeting similar pathways have been grouped by color. (B) Transverse cross section of the trunk of an embryo during neural crest migration. Signaling requirements for ventral neural crest migration and sympathetic specification are indicated by bold text. Signaling cues targeted by the protocols in (A) are indicated with colored text matching those in the figure legend. (C) Schematic view of the signaling requirements for sympathetic precursor proliferation and target innervation. The discontinuous axon and blood vessel represent the large distance from the sympathetic ganglia to their peripheral targets. * Aphidicolin selection yields optimal cell purity. However, this is not required. AA, ascorbic acid; BDNF, brain-derived neurotrophic factor; BMP, bone morphogenetic protein; CHIR, CHIR99021; CXCL12, C-X-C motif chemokine ligand 12; dbcAMP, dibutyryl cyclic adenosine monophosphate; EGF, epidermal growth factor; eGFP, enhanced green fluorescent protein; FGF, fibroblast growth factor; FSK, forskolin; GDNF, glial cell line derived neurotrophic factor; IGF, insulin-like growth factor; MYCN, MYCN proto-oncogene; NGF, nerve growth factor; NRG1, neuregulin 1; NRTN, neurturin; NTF3, neurotrophin 3; PHOX2B, paired-like homeobox 2b; PMP, purmorphamine; RA, retinoic acid; SEMA3A, semaphorin 3A; SB, SB431542; SDIA, stromal cell-derived-inducing activity; SHH, Sonic hedgehog; TGFβ, transforming growth factor beta.
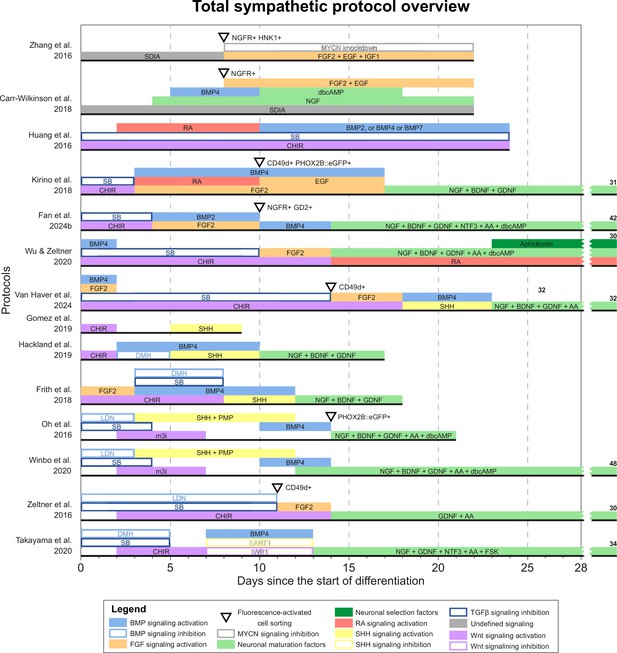
Total overview of all sympathetic protocols.
Overview of all unique sympathetic protocols. Protocol durations are indicated by the horizontal black bars. Total protocol durations exceeding the width of the graph are indicated to the right of the graph. Molecules targeting similar pathways have been grouped by color. AA, ascorbic acid; BDNF, brain-derived neurotrophic factor; BMP, bone morphogenetic protein; CD49d, integrin subunit α4; CHIR, CHIR99021; dbcAMP, dibutyryl cyclic adenosine monophosphate; DMH, dorsomorphin; EGF, epidermal growth factor; eGFP, enhanced green fluorescent protein; FGF, fibroblast growth factor; FSK, forskolin; GD2, disialoganglioside; GDNF, glial cell line-derived neurotrophic factor; HNK1, human natural killer-1; IGF1, insulin-like growth factor 1; LDN, LDN193189; m3i, modified three inhibitor approach (CHIR99021, DAPT, and PD173074); MYCN, MYCN proto-oncogene; NGF, nerve growth factor; NGFR, nerve growth factor receptor; NTF3, neurotrophin 3; PHOX2B, paired-like homeobox 2B; PMP, purmorphamine; RA, retinoic acid; SB, SB431542; SDIA, stromal cell-derived inducing activity; SHH, Sonic hedgehog; TGFβ, transforming growth factor.
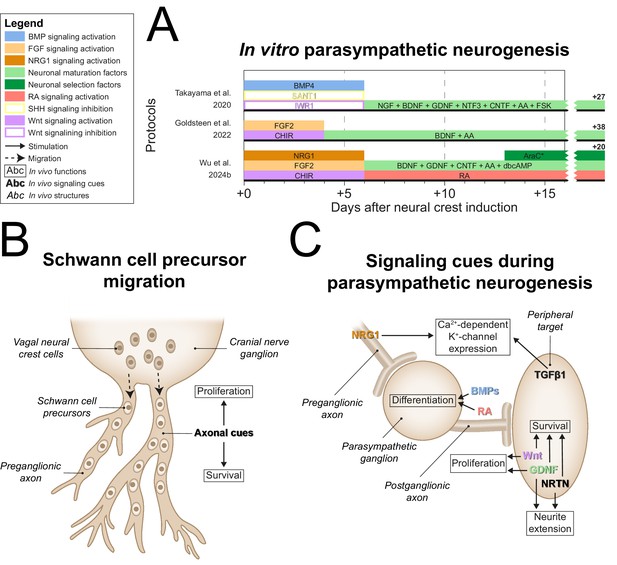
Parasympathetic neurogenesis.
(A) Timings and signaling cues used from neural crest induction until the end of parasympathetic neuron differentiation. Duration of this phase per protocol is indicated by the horizontal black bars. Total duration of this phase per protocol is indicated to the right of the graph. Molecules targeting similar pathways have been grouped by color. (B) Migration of vagal neural crest-derived Schwann cell precursors along a cranial nerve. (C) Schematic view of the signaling requirements for parasympathetic precursor proliferation and target innervation. Signaling requirements are indicated by bold text. Signaling cues targeted by the protocols in (A) are indicated with colored text matching those in the figure legend. * Cytosine arabinoside selection yields optimal cell purity. However, this is not required. AA, ascorbic acid; AraC, cytosine arabinoside; BDNF, brain-derived neurotrophic factor; BMP, bone morphogenetic protein; CHIR, CHIR99021; CNTF, ciliary neurotrophic factor; dbcAMP, dibutyryl cyclic adenosine monophosphate; FGF, fibroblast growth factor; FSK, forskolin; GDNF, glial cell line-derived neurotrophic factor; NGF, nerve growth factor; NRG1, neuregulin 1; NRTN, neurturin; NTF3, neurotrophin 3; RA, retinoic acid; SHH, Sonic hedgehog; TGFβ, transforming growth factor beta.
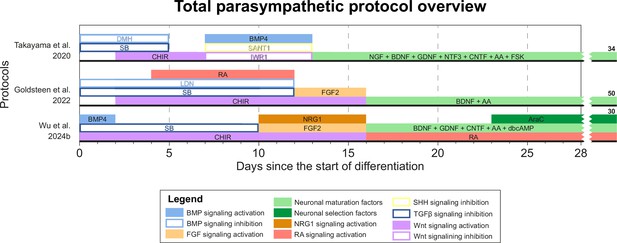
Total overview of all parasympathetic protocols.
Overview of all unique parasympathetic protocols. Protocol durations are indicated by the horizontal black bars. Total protocol durations are indicated to the right of the graph. Molecules targeting similar pathways have been grouped by color. AA, ascorbic acid; AraC, cytosine arabinoside; BDNF, brain-derived neurotrophic factor; BMP, bone morphogenetic protein; CHIR, CHIR99021; CNTF, ciliary neurotrophic factor; dbcAMP, dibutyryl cyclic adenosine monophosphate; DMH, dorsomorphin; FGF, fibroblast growth factor; FSK, forskolin; GDNF, glial cell line-derived neurotrophic factor; LDN, LDN193189; NGF, nerve growth factor; NRG1, neuregulin 1; NTF3, neurotrophin 3; RA, retinoic acid; SB, SB431542; SHH, Sonic hedgehog; TGFβ, transforming growth factor.
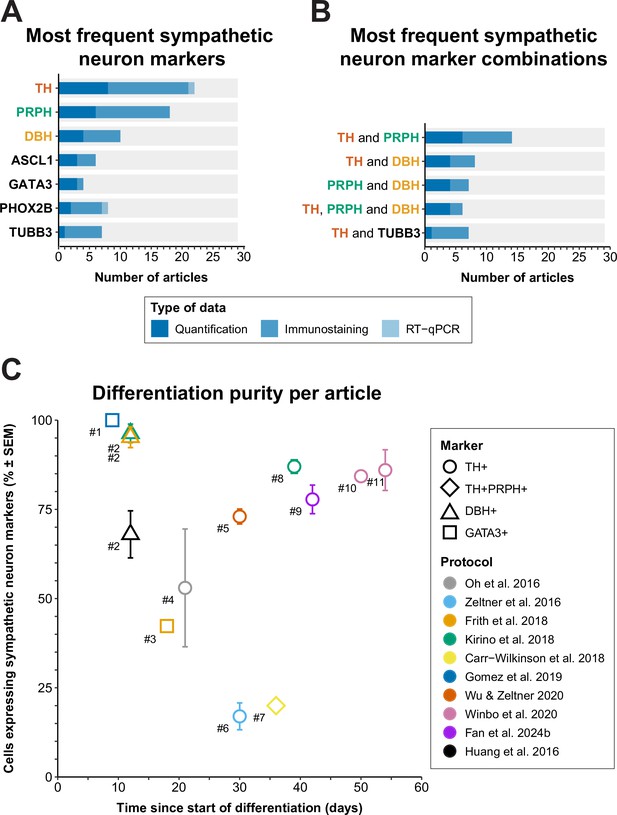
Sympathetic neuron definitions and differentiation efficiency.
(A) All sympathetic neuron markers used in ≥3 articles, stratified by technique. (B) All combinations of sympathetic neuron markers used in ≥6 articles. Markers featured in multiple combinations are marked by colored text in (A) and (B). (C) Scatter plot of protocol purity and time of quantification per article. The graph shows only the latest timepoint per article %TH+ (or %GATA3 + or %DBH+, if %TH+ was not determined) was measured. Shapes indicate the markers used for quantification and protocol applied per article is indicated by color. #1 Gomez et al., 2019, #2 Cheng et al., 2024, #3 Frith et al., 2018, #4 Oh et al., 2016, #5 Wu et al., 2022b, #6 Zeltner et al., 2016, #7 Carr-Wilkinson et al., 2018, #8 Kirino et al., 2018, #9 Fan et al., 2024a, #10 Li et al., 2023, #11 Winbo et al., 2020. Sample sizes per article can be found in Source data 1. ASCL1, achaete-scute family bHLH transcription factor 1; DBH, dopamine beta-hydroxylase; GATA3, GATA binding protein 3; RT-qPCR, quantitative reverse transcriptase polymerase chain reaction; PHOX2B, paired-like homeobox 2B; PRPH, peripherin; SEM, standard error of the mean; TH, tyrosine hydroxylase; TUBB3, tubulin beta 3 class III.
Tables
Characteristics of the included articles.
Articles are grouped per protocol and neuron type, in chronological order. Rows in bold indicate the earliest article per protocol.
| Reference | Journal (ISO 4) | Neuron type | Article type | Source cells |
|---|---|---|---|---|
| Huang et al., 2016 | Sci Rep | Sympathetic | Protocol development | PSCs |
| Cheng et al., 2024* | Sci Rep | Sympathetic | Protocol application | iPSCs |
| Zhang et al., 2016 | PLoS One | Sympathetic | Protocol development/application | ESCs |
| Oh et al., 2016 | Cell Stem Cell | Sympathetic | Protocol development | PSCs |
| Zeltner et al., 2016 | Nat Med | Sympathetic | Protocol development/application | PSCs |
| Saito-Diaz et al., 2019 | Curr Protoc Stem Cell Biol | Sympathetic | Methodological | PSCs |
| Frith et al., 2018 | eLife | Sympathetic | Protocol development | PSCs |
| Frith and Tsakiridis, 2019 | Curr Protoc Stem Cell Biol | Sympathetic | Methodological | PSCs |
| Saldana-Guerrero et al., 2024 | Nat Commun | Sympathetic | Protocol application | ESCs |
| Kirino et al., 2018 | Sci Rep | Sympathetic | Protocol development | PSCs |
| Amer-Sarsour et al., 2024 | Embo J | Sympathetic | Protocol application | iPSCs |
| Saleh et al., 2024 | Free Radic Biol Med | Sympathetic | Protocol application | iPSCs |
| Carr-Wilkinson et al., 2018 | Stem Cells Int | Sympathetic | Protocol development | ESCs |
| Hackland et al., 2019 | Stem Cell Reports | Sympathetic | Protocol development/application | PSCs |
| Gomez et al., 2019 | Development | Sympathetic | Protocol development/application | PSCs |
| Wu and Zeltner, 2020 | J Vis Exp | Sympathetic | Methodological | ESCs |
| Wu et al., 2022a | Clin Auton Res | Sympathetic | Protocol application | PSCs |
| Wu et al., 2022b | Nat Commun | Sympathetic | Protocol development/application | PSCs |
| Wu et al., 2023 | Front Neurosci | Sympathetic | Protocol application | ESCs |
| Wu et al., 2024a | STAR Protoc | Sympathetic | Methodological | ESCs |
| Winbo et al., 2020 | Am J Physiol Heart Circ Physiol | Sympathetic | Protocol development | iPSCs |
| Winbo et al., 2021 | Am J Physiol Heart Circ Physiol | Sympathetic | Protocol application | iPSCs |
| Bernardin et al., 2022 | Cells | Sympathetic | Protocol application | iPSCs |
| Li et al., 2023 | Philos Trans R Soc Lond B Biol Sci | Sympathetic | Protocol application | iPSCs |
| Van Haver et al., 2024 | iScience | Sympathetic | Protocol development/application | PSCs |
| Fan et al., 2024a | J Mol Neurosci | Sympathetic | Protocol development | PSCs |
| Fan et al., 2024b | Cell Rep Med | Sympathetic | Protocol application | PSCs |
| Takayama et al., 2020 | Sci Rep | Sympathetic or parasympathetic | Protocol development | PSCs |
| Takayama et al., 2023 | Int J Mol Sci | Sympathetic or parasympathetic | Protocol application | PSCs |
| Akagi et al., 2024a | FEBS Open Bio | Parasympathetic | Protocol application | iPSCs |
| Akagi et al., 2024b | Molecules | Sympathetic or parasympathetic | Protocol application | iPSCs |
| Goldsteen et al., 2022 | Front Pharmacol | Parasympathetic | Protocol development | ESCs |
| Wu et al., 2024b | Cell Stem Cell | Parasympathetic | Protocol development/application | PSCs |
-
ESCs, embryonic stem cells; iPSCs, induced pluripotent stem cells; ISO, International Organization for Standardization; PSCs, pluripotent stem cells (iPSCs or ESCs).
-
*
Cheng et al., 2024 applied three of the protocols included in this review, by Huang et al., 2016, Frith et al., 2018, and Kirino et al., 2018.
Patch clamp recordings of hPSC-derived sympathetic neurons.
Electrophysiological characteristics of hPSC-derived sympathetic neurons determined by whole-cell patch clamp. Data from primary adult murine thoracic sympathetic neurons is included for reference. Tabulation is in chronological order. Data is reported as mean ± SEM or range, unless indicated otherwise. AP, action potential; hPSC, human pluripotent stem cell; NR, not reported; SEM, standard error of the mean.
| Adult murine thoracic sympathetic neurons (McKinnon et al., 2019) (n=35) | Oh et al., 2016 (n=9) | Frith et al., 2018 (n=14) | Winbo et al., 2020 (n=30) | Takayama et al., 2023 (n=113) | |
|---|---|---|---|---|---|
| Age (days) | 37–379 (postnatal) | 28 | >20 | 48–76 | >42 |
| Membrane capacitance (pF) | 89 ± 4.6 (n=34) | NR | 11 ± 0.6 | 85 ± 5.1 | NR |
| Current injection range (pA) | 0–200 | 0–800 | –10 to 100 | 0–300 | –100 to 300 |
| Proportion neurons firing repetitive APs, % | 100 | 56 | 21 | 73 | 36 |
| Resting membrane potential (mV) | −60 ± 1.1 | −46 ± 5.4 | −54 to –60 | −60 ± 1.9 | NR |
| AP amplitude (mV) | 54 ± 2.7 | NR | NR | 93 ± 3.9 | 74 ± 4.3 (n=20) |
| AP duration, half-width (ms) | 4.6 ± 0.2 | NR | NR | 2.8 ± 0.2 | NR |
Additional files
-
Supplementary file 1
List of excluded articles.
A list of all articles that were excluded during full-text screening, including the reason of exclusion. PSCs, pluripotent stem cells.
- https://cdn.elifesciences.org/articles/103728/elife-103728-supp1-v2.docx
-
Supplementary file 2
Quality assessment criteria and definitions.
Detailed criteria and definitions per judgement per criterion.
- https://cdn.elifesciences.org/articles/103728/elife-103728-supp2-v2.docx
-
Supplementary file 3
Methodological details of included protocols.
- https://cdn.elifesciences.org/articles/103728/elife-103728-supp3-v2.docx
-
Supplementary file 4
Small molecules used in autonomic neuron protocols.
Modes of actions for all small molecules featured in this article.
- https://cdn.elifesciences.org/articles/103728/elife-103728-supp4-v2.docx
-
Supplementary file 5
Molecular autonomic neuron markers.
Function and expression profiles of molecular autonomic neuron markers.
- https://cdn.elifesciences.org/articles/103728/elife-103728-supp5-v2.docx
-
Supplementary file 6
Preferred Reporting Items for Systematic Reviews and Meta-Analyses 2020 Main Checklist.
Locations of all Preferred Reporting Items for Systematic Reviews and Meta-Analyses 2020 Main Checklist items.
- https://cdn.elifesciences.org/articles/103728/elife-103728-supp6-v2.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/103728/elife-103728-mdarchecklist1-v2.docx
-
Source data 1
Dataset relating to Figure 1.
All title-abstract exclusion reasons per record. Please see Supplementary file 1 for exclusion reasons during full-text screening. n/a, not available. Dataset relating to Figure 7A and B. Definitions of sympathetic neurons per article. Only one of quantification, immunofluorescence or RT-qPCR data was collected per article. Quantification data was preferentially collected over immunofluorescence or RT-qPCR. Immunofluorescence data was collected preferentially over RT-qPCR. n/a, not available; RT-qPCR, quantitative reverse transcriptase polymerase chain reaction. Dataset relating to Figure 7C: sympathetic neuron efficiency quantification per article. Only one efficiency marker was collected per article. %TH+ was collected preferentially over the other markers. %TH+ PRPH+ was collected preferentially over %GATA3+ and %DBH+. DBH, dopamine beta-hydroxylase; GATA3, GATA binding protein 3; n/a, not available; NR, not reported; PRPH, peripherin; TH, tyrosine hydroxylase.
- https://cdn.elifesciences.org/articles/103728/elife-103728-data1-v2.xlsx





