Numb provides a fail-safe mechanism for intestinal stem cell self-renewal in adult Drosophila midgut
Figures
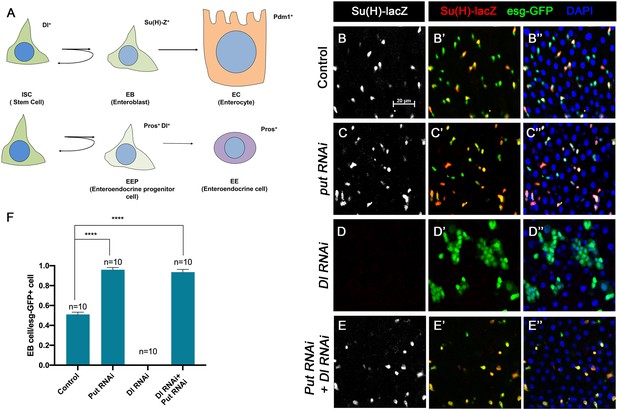
BMP signaling inhibits Dl-independent N pathway activity to promote intestinal stem cell (ISC) self-renewal.
(A) A scheme for the ISC lineage in Drosophila midgut. (B-E’’) Representative images of Control guts (B–B’’), midguts expressing UAS-Put-RNAi (C–C’’), UAS-Dl-RNAi (D–D’’), or UAS-Put-RNAi+UAS-Dl-RNAi (E–E’’) with esg-Gal4ts, UAS-GFP at 29°C for 10 days and immunostained for Su(H)-lacZ (gray or red) and GFP (green). Su(H)-lacZ is used as a marker for enteroblast (EB). DAPI (blue) staining indicates nuclei. Compared with control guts (B–B’’), Put knockdown (C–C’’) in precursor cells (green) caused an increase of EB pairs. Dl knockdown induced stem cell-like tumor. Put and Dl double knockdown induced a dramatic increase of EBs. (F) Quantification of percentage of EB cells of each genotype. Data are mean ± SD from three independent experiments. *p<0.05, ****p < 0.0001. One-way ANOVA was performed for statistical comparisons. Scale bar (20 μm) is shown in B.
-
Figure 1—source data 1
Source data for the quantification in Figure 1.
- https://cdn.elifesciences.org/articles/104723/elife-104723-fig1-data1-v1.xlsx
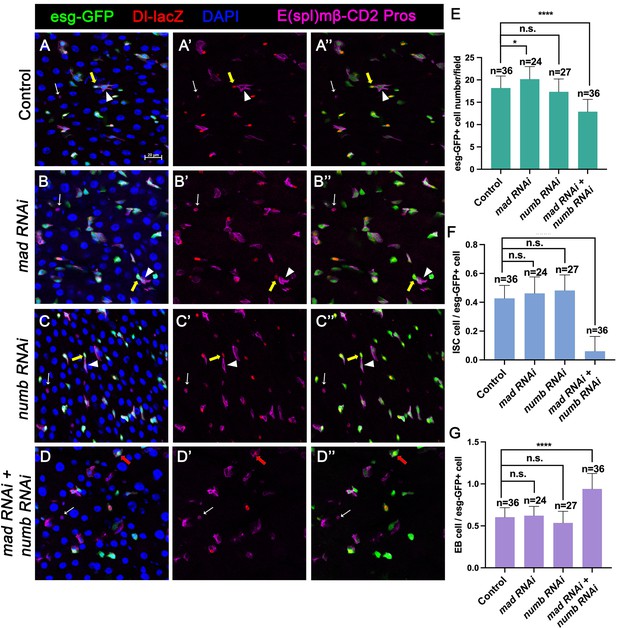
Numb is important for intestinal stem cell (ISC) maintenance when BMP pathway activity is attenuated.
(A-D’’) Representative images of adult midguts expressing UAS-mCherry-RNAi (Control) (A–A’’), UAS-Mad-RNAi (B–B’’), UAS-Numb-RNAi (C–C’’), and UAS-Mad-RNAi+UAS-Numb-RNAi (D–D’’) with esg-Gal4ts, UAS-GFP at 30°C for 14 days and immunostained for Dl-lacZ (red), E(spl)mβ-CD2 (cytoplasmic magenta), and Pros (nuclear magenta), which are markers for ISC, enteroblast (EB), and enteroendocrine (EE), respectively. DAPI (blue) staining indicates nuclei. Yellow arrows indicate ISCs (Dl-lacZ+ E(spl)mβ-CD2− Pros−), white arrowheads indicate EBs (E(spl)mβ-CD2+), and white arrows indicate EEs (Pros+) in Control, Mad, or Numb single knockdown guts. Red arrow indicated a Dl-lacZ+, E(spl) mβ-CD2+ cells in Mad and Numb double knockdown guts. Scale bar (20 μm) is presented in (A). (E–G) Quantification of number of precursor cells (E), percentage of ISC cells (F), and percentage of EB cells (G) of each genotype. Data are mean ± SD from three independent experiments. *p < 0.05, ****p < 0.0001. One-way ANOVA was performed for statistical comparisons.
-
Figure 2—source data 1
Source data for the quantification in Figure 2E–G.
- https://cdn.elifesciences.org/articles/104723/elife-104723-fig2-data1-v1.xlsx
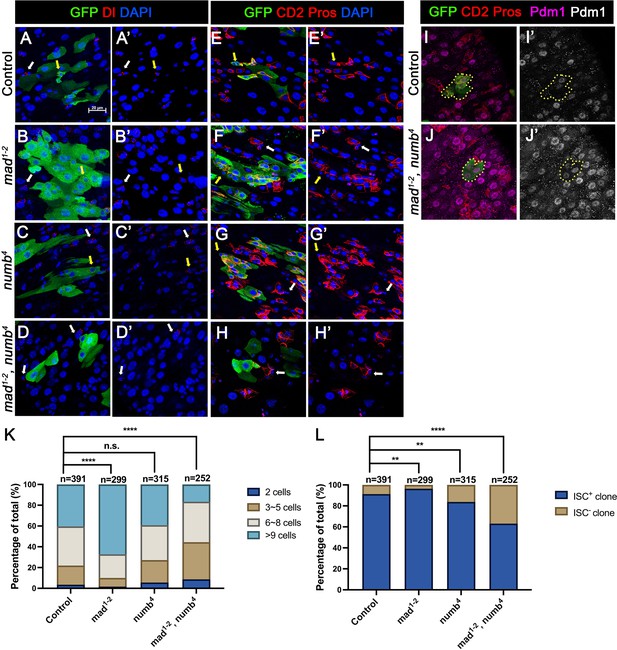
numb and mad double mutations resulted in loss of intestinal stem cell (ISC).
(A-H’) Representative images of adult midguts containing MARCM clones (green) of FRT40 (Control) (A, A’, E, E’), mad1-2 (B, B’, F, F’), numb4 (C, C’, G, G’), and mad1-2, numb4 (D, D’, H, H’) and immunostained for GFP (green) and Dl (red in A–D’) or E(spl)mβ-CD2 and Pros (red in E–H’) at 14 days (grown at 18°C) after clone induction. GFP marks the clones. DAPI (blue) staining indicates nuclei. ISCs inside and outside the clones are indicated by yellow and white arrows, respectively. (I) Representative images of adult midguts containing MARCM clones (green) of control (I, I’) or mad1-2, numb4 (J, J’) immunostained for GFP (green), E(spl)mβ-CD2 and Pros (red), and Pdm1 (magenta and gray). Scale bar (20 μm) is presented in (A). (K) Quantification of clone size for the indicated genotypes 14 days after clone induction. (L) Quantification of numbers of clones with or without ISCs. Data are mean ± SD from three independent experiments. **p < 0.01, ****p < 0.0001. ᵡ2 test was performed for statistical comparisons.
-
Figure 3—source data 1
Source data for the quantification in Figure 3L.
- https://cdn.elifesciences.org/articles/104723/elife-104723-fig3-data1-v1.xlsx
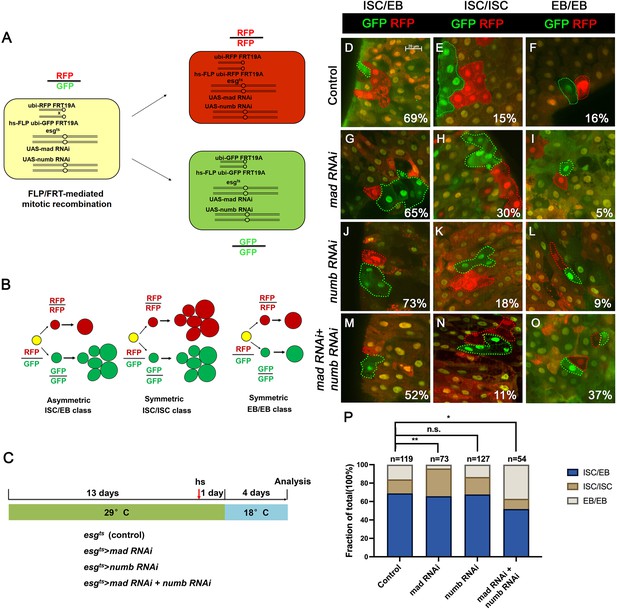
Depletion of both Numb and Mad leads to more symmetric intestinal stem cell (ISC) divisions that produce two enteroblasts (EBs).
(A) Scheme of an ISC division that produces differentially labeled daughter cells (RFP+ GFP− and RFP− GFP+) through FRT-mediated mitotic recombination. Adapted from Tian and Jiang, 2014. (B) Scheme of differentially labeled twin clones generated by FLP/FRT-mediated mitotic recombination of dividing ISCs. Adapted from Tian and Jiang, 2014. (C) Scheme of twin-spot experiments. Three- to five-day-old adult flies of indicated genotype are grown at 29°C for 14 days before heat shock to induce clones. After 1-day recovery at 29°C, the flies are raised at 18°C for 4 days prior to analysis. (D–O) Representative images of twin-spot clones from adult midguts of the indicated genotypes. Scale bar 20 μm is shown in (D). (P) Quantification of twin spots of different classes from guts of the indicated genotypes. Data are mean ± SD from three independent experiments. *p < 0.05, **p < 0.01. ᵡ2 test was performed for statistical comparisons.
-
Figure 4—source data 1
Source data for the quantification in Figure 4P.
- https://cdn.elifesciences.org/articles/104723/elife-104723-fig4-data1-v1.xlsx
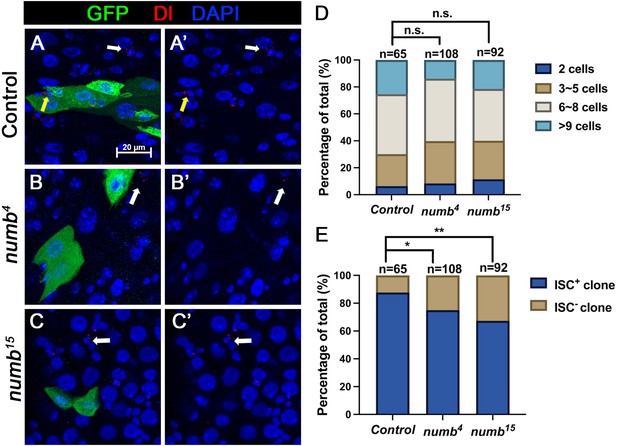
numb mutant clones exhibit weak stem cell loss phenotype.
(A-C’) Representative images of adult midguts containing MARCM clone (green) of FRT40 (Control) (A, A’), numb4 (B, B’), and numb15 (C, C’) and immunostained for Dl (red), GFP (green), and DAPI (blue) at 14 days after clone induction. GFP marks the clones. Intestinal stem cells (ISCs) inside and outside the clones are indicated by yellow and white arrows, respectively. Scale bar (20 μm) is shown in (A). (D) Quantification of clone size distribution for the indicated genotypes at 14 days after clone induction. (E) Quantification of numbers of clones with or without ISC. Data are mean ± SD from three independent experiments. *p < 0.05, **p < 0.01. ᵡ2 test was performed for statistical comparisons.
-
Figure 5—source data 1
Source data for the quantification in Figure 5E.
- https://cdn.elifesciences.org/articles/104723/elife-104723-fig5-data1-v1.xlsx
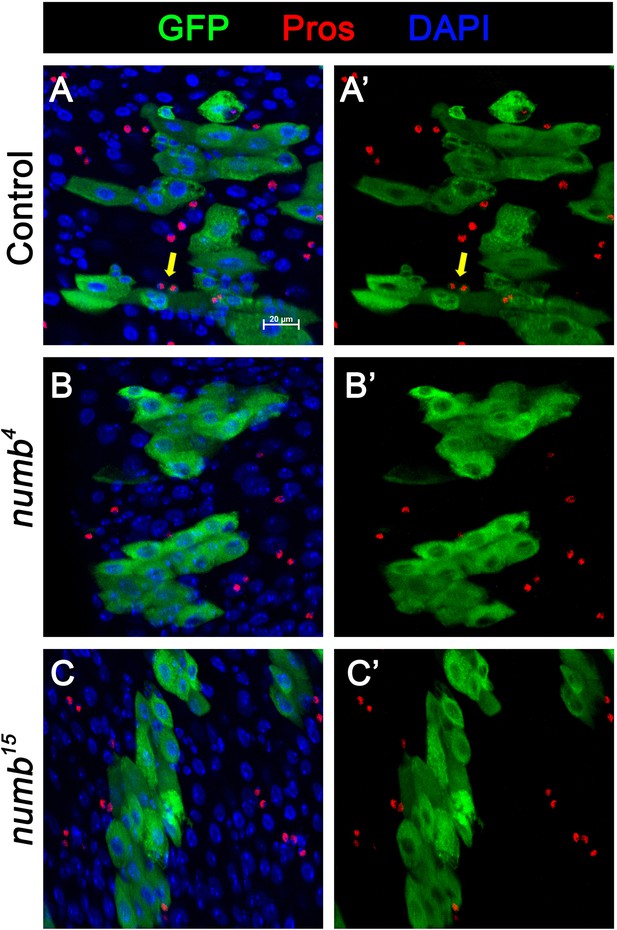
Numb is required for enteroendocrine (EE) fate determination.
(A-C’) MARCM clone (green) of control (A, A’), numb4 (B, B’), and numb15 (C, C’) are stained for Pros (red) at 14 days after clone induction. Representative clone in control guts (A, A’) contains EE cells (Pros positive), as indicated with yellow arrows. Representative clones in numb4 (B, B’) and numb15 (C, C’) guts do not contain any EE cells. Scale bar (20 μm) is presented in (A).
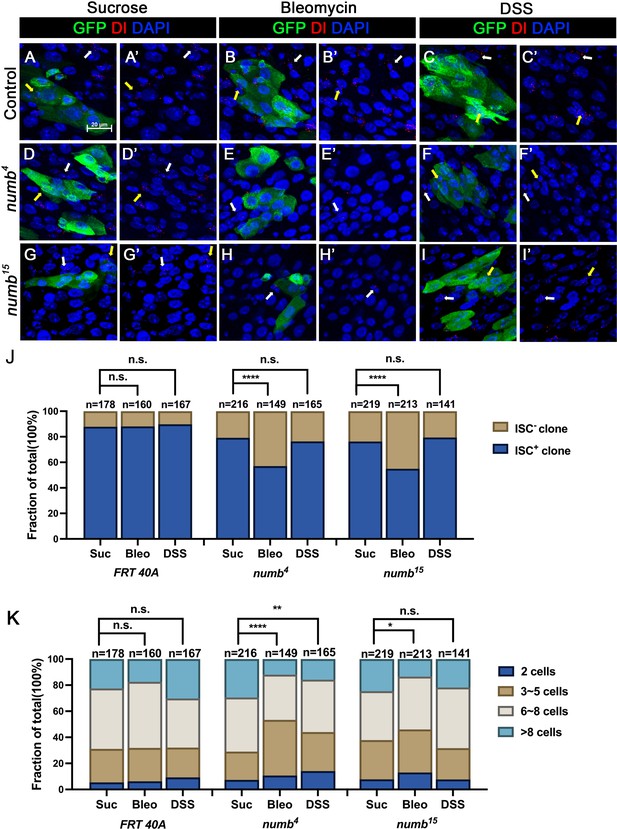
Numb is critical for intestinal stem cell (ISC) maintenance during regeneration.
(A–I’) Adult flies of indicated genotype were treated with sucrose, bleomycin, or dextran sodium sulfate (DSS) for 24 hr at 14 days after clone induction and recovered for another 4 days before dissection. Guts containing MARCM clones of the indicated genotype were stained for GFP (green) and Dl (red). GFP marks the clones. DAPI (blue) staining indicates the nuclei. Stem cells inside and outside the clones are indicated by yellow and white arrows, respectively. Scale bar (20 μm) is shown in (A). (J) Quantification of the percentage of clones with or without ISCs. (K) Quantification of clone size distribution for the indicated genotypes. Data are mean ± SD from three independent experiments. *p < 0.05, **p < 0.01, ****p < 0.0001. ᵡ2 test was performed for statistical comparisons.
-
Figure 6—source data 1
Source data for the quantification in Figure 6J.
- https://cdn.elifesciences.org/articles/104723/elife-104723-fig6-data1-v1.xlsx
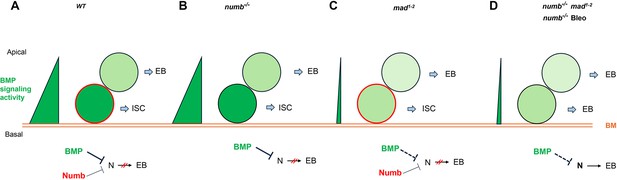
Model for Numb and BMP signaling in intestinal stem cell (ISC)/enteroblast (EB) fate decision.
(A) During asymmetric ISC division, the basal ISC daughter transduces higher level of BMP signaling and inherits higher level of Numb activity than the apical one. Inhibition of N by BMP signaling and Numb promotes ISC fate. (B) In numb mutant background, differential BMP signaling between the basal and apical ISC daughters is sufficient to generate differential N pathway activities to drive asymmetric fate decision. (C) In mad mutant background, the shallow BMP activity gradient acts in conjunction with the asymmetric Numb activity to generate differential N pathway activities between the basal and apical ISC daughters to drive asymmetric fate decision. (D) In numb mad double mutant background or in guts containing numb mutant clones and injured by bleomycin (Bleo) feeding, the shallow BMP activity gradient is often insufficient to generate asymmetric N pathway activation, leading to precocious ISC-to-EB differentiation. BM: basement membrane; Bleo: bleomycin; thin and dashed lines indicate weak inhibition.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (D. melanogaster) | esg-Gal4 | Jiang et al., 2009 | FLYB: FBti0013268 | FlyBase symbol: P{GawB}NP5130 |
| Genetic reagent (D. melanogaster) | Dl-lacZ | Zeng et al., 2010 | FLYB: FBti0004778 | FlyBase symbol: P{PZ}Delta[05151] |
| Genetic reagent (D. melanogaster) | Su(H)-lacZ | Zeng et al., 2010 | FLYB: FBtp0014034 | FlyBase symbol: P{Ddc.E(spl)m8-HLH-lacZ.Gbe} |
| Genetic reagent (D. melanogaster) | E(spl)mβCD2 | Bloomington Drosophila Stock Center | BDSC 83353 FLYB: FBst0083353 RRID:BDSC_83353 | FlyBase symbol: w[*]; l(2)*[*]/CyO, P{ry[+t7.2]=en1}wg[en11]; P{w[+mC]=E(spl)mbeta-HLH-CD2.dC}T6 |
| Genetic reagent (D. melanogaster) | Tub-Gal80ts | Jiang et al., 2009 | FLYB: FBti0027796 | FlyBase symbol: P{tubP-GAL80ts} |
| Genetic reagent (D. melanogaster) | UAS-Put RNAi | Vienna Drosophila Resource Center | VDRC 107071 FLYB: FBst0478894 RRID:Flybase_FBst0473060 | FlyBase symbol: P{KK102676}VIE-260B |
| Genetic reagent (D. melanogaster) | UAS-Dl RNAi | Bloomington Drosophila Stock Center | BDSC 28032 FLYB: FBst0028032 RRID:BDSC_28032 | FlyBase symbol: y (1) v(1); P{TRiP.JF02867}attP2 |
| Genetic reagent (D. melanogaster) | UAS-Mad RNAi | Vienna Drosophila Resource Center | VDRC 12635 FLYB: FBst0450590 RRID:Flybase_FBst0450590 | FlyBase symbol: w1118; P{GD4121}v12635 |
| Genetic reagent (D. melanogaster) | UAS-Numb RANi | Bloomington Drosophila Stock Center | BDSC 35045 FLYB: FBst0035045 RRID:BDSC_35045 | FlyBase symbol: y(1) sc[*] v(1) sev(21); P{y[+t7.7] v[+t1.8]=TRiP.HMS01459}attP2 |
| Genetic reagent (D. melanogaster) | UAS-mCherry RANi | Bloomington Drosophila Stock Center | BDSC 35785 FLYB: FBst0035785 RRID:BDSC_35785 | FlyBase symbol: y(1) sc[*] v(1) sev(21); P{y[+t7.7] v[+t1.8]=VALIUM20-mCherry.RNAi}attP2 |
| Genetic reagent (D. melanogaster) | numb4 | Skeath and Doe, 1998 | FLYB: FBal0090215 | FlyBase symbol: numb(4) |
| Genetic reagent (D. melanogaster) | numb15 | Sallé et al., 2017 | FLYB: FBal0146969 | FlyBase symbol: numb(15) |
| Genetic reagent (D. melanogaster) | mad1-2 | Bloomington Drosophila Stock Center | BDSC 7323 FLYB: FBst0007323 RRID:BDSC_7323 | FlyBase symbol: w*; Mad(; 1-2) P{neoFRT}40A/CyO |
| Genetic reagent (D. melanogaster) | FRT 40A | Bloomington Drosophila Stock Center | BDSC 8212 FLYB: FBst0008212 RRID:BDSC_8212 | FlyBase symbol: w[1118]; P{ry[+t7.2]=neoFRT}40A/CyO; P{ry[+t7.2]=ey-FLP.N}6, ry[506] |
| Genetic reagent (D. melanogaster) | yw, hs-FLP | Bloomington Drosophila Stock Center | BDSC 1929 FLYB: FBst0001929 RRID:BDSC_1929 | FlyBase symbol: P{ry[+t7.2]=hsFLP}12, y(1) w[*]; sna[Sco]/CyO |
| Genetic reagent (D. melanogaster) | FRT 40A, tub-GAL80 | Bloomington Drosophila Stock Center | BDSC 5192 FLYB: FBst0005192 RRID:BDSC_5192 | FlyBase symbol: y(1) w[*]; P{w[+mC]=tubP-GAL80}LL10 P{ry[+t7.2]=neoFRT}40A/CyO |
| Genetic reagent (D. melanogaster) | UAS-GFP | Bloomington Drosophila Stock Center | BDSC 5130 FLYB: FBst0005130 RRID:BDSC_5130 | FlyBase symbol: y(1) w[*]; betaTub60D[Pin-Yt]/CyO; P{w[+mC]=UAS-mCD8::GFP.L}LL6 |
| Genetic reagent (D. melanogaster) | FRT 19A | Bloomington Drosophila Stock Center | BDSC 1709 FLYB: FBst0001709 RRID:BDSC_1709 | FlyBase symbol: P{ry[+t7.2]=neoFRT}19A; ry[506] |
| Genetic reagent (D. melanogaster) | Ubi-GFPnls | Chen and Schüpbach, 2006 | FLYB: FBti0015575 | FlyBase symbol: P{Ubi-GFP(S65T)nls}X |
| Genetic reagent (D. melanogaster) | FRT 19A, ubi-mRFPnls | Bloomington Drosophila Stock Center | BDSC 31418 FLYB: FBst0031418 RRID:BDSC_31418 | FlyBase symbol: P{w[+mC]=Ubi-mRFP.nls}1, w[*], P{ry[+t7.2]=hsFLP}12P{ry[+t7.2]=neoFRT}19A |
| Antibody | anti-GFP (chicken polyclonal) | Abcam | Cat#: ab13970; RRID:AB_300798 | IF (1:1000) |
| Antibody | anti-β-Galactosidase (rabbit polyclonal) | MP Biomedicals | Cat#: 08559761 RRID:AB_3675281 | IF (1:1000) |
| Antibody | anti-Rat CD2 (mouse monoclonal) | Bio-Rad | Cat#: MCA154GA RRID:AB_566608 | IF (1:2000) (Formerly AbD Serotec) |
| Antibody | anti-Dl extracellular domain (mouse monoclonal) | DSHB | Cat#: c594.9b RRID:AB_528194 | IF (1:20) |
| Antibody | anti-Prospero (mouse monoclonal) | DSHB | Cat#: Prospero RRID:AB_528440 | IF (1:20) |
| Antibody | anti-Pdm1 (rabbit polyclonal) | Dr. Xiaohang Yang | IF (1:1000) | |
| Antibody | anti-Chicken Alexa Fluor 488 (goat polyclonal secondary) | Thermo Fisher Scientific | Cat#: A-11039 RRID:AB_2534096 | IF (1:1000) |
| Antibody | anti-Mouse Alexa Flour 546 (goat polyclonal secondary) | Thermo Fisher Scientific | Cat#: A-11030 RRID:AB_2737024 | IF (1:1000) |
| Antibody | anti-Rabbit Alexa Flour 546 (goat polyclonal secondary) | Thermo Fisher Scientific | Cat#: A-11035 RRID:AB_2534093 | IF (1:1000) |
| Antibody | anti-Mouse Cy5 (goat polyclonal) | Jackson ImmunoResearch Labs | Cat#: 115-175-166 RRID:AB_2338714 | IF (1:500) |
| Antibody | anti-Rabbit Cy5 (goat polyclonal) | Jackson ImmunoResearch Labs | Cat#: 111-175-144 RRID:AB_2338013 | IF (1:500) |
| Chemical compound, drug | DAPI (4′,6-diamidino-2-phenylindole, dihydrochloride) | Invitrogen | Cat#: D1306 | IF (1:2000) |
| Chemical compound, drug | DSS (dextran sulfate sodium) | Sigma-Aldrich | Cat#: 42867 | 5% solution |
| Chemical compound, drug | Bleomycin sulfate from Streptomyces verticillus | Sigma-Aldrich | Cat#: B8416 | 25 μg/ml |
| Chemical compound, drug | Sucrose | Fisher Bioreagent | Cat#: BP220-212 | 5% solution |





