N6-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression
Figures
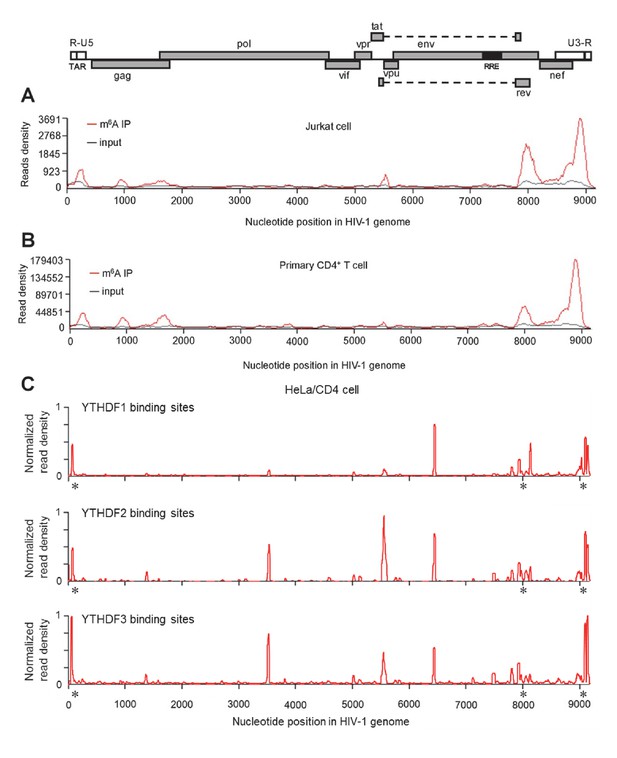
HIV-1 RNA contains m6A modifications and YTHDF1–3 proteins bind to m6A-modified HIV-1 RNA.
(A–B) The distribution of m6A reads from m6A-seq mapped to HIV-1 genome (red line) in HIV-1 infected Jurkat cells (A) or primary CD4+ T-cells (B). Baseline signal from the RNA-seq of input samples is shown as a black line. A schematic diagram of HIV-1NL4-3 genome is shown above. TAR, transacting response element; RRE, Rev response element. Jurkat cells (A) or primary CD4+ T-cells (B) were infected with HIV-1NL4-3 and total RNA was extracted for m6A-seq at 72 or 96 hr post-infection (hpi), respectively. (C) YTHDF1-3 proteins bind to the HIV-1 gRNA. HeLa/CD4 cells overexpressing FLAG-tagged YTHDF1-3 proteins were infected with HIV-1NL4-3 (MOI= 5) for 72 hr and used in CLIP-seq assay to identify their binding sites on HIV-1 gRNA. The distribution of mapped reads (>16 nt) with corresponding nucleotide positions are shown, forming peaks as putative binding positions. Asterisks mark the peak clusters overlapping with identified m6A peaks, indicating high-confident YTHDFs binding sites. Read density was normalized to the total number of mapped reads in each sample (YTHDF1: 28438; YTHDF2: 232568; YHTDF3: 124915). The data presented are representative of results from two independent experiments (n=2).
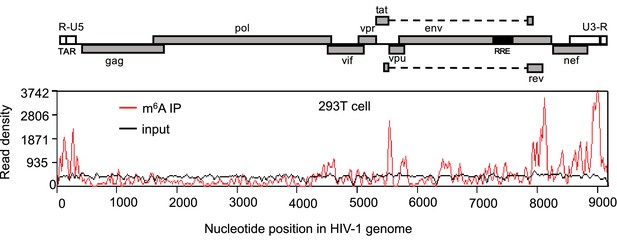
HIV-1 RNA contains m6A modifications.
HEK293 T cells were transfected with a proviral DNA-containing plasmid (pNL4-3). Total RNA was extracted at 48 hr post-transfection and immunoprecipitated with an m6A-specific antibody. Enriched RNA was subjected to next generation sequencing. Peaks show the relative abundance of m6A sites on the HIV-1 genome. The distribution of m6A reads from m6A-seq mapped to HIV-1 genome (red line). Baseline signal from the RNA-seq of input samples is shown as a black line. A schematic diagram of HIV-1NL4-3 genome features is shown above. TAR, transacting response element; RRE, Rev response element. The data presented are representative of two independent experiments.
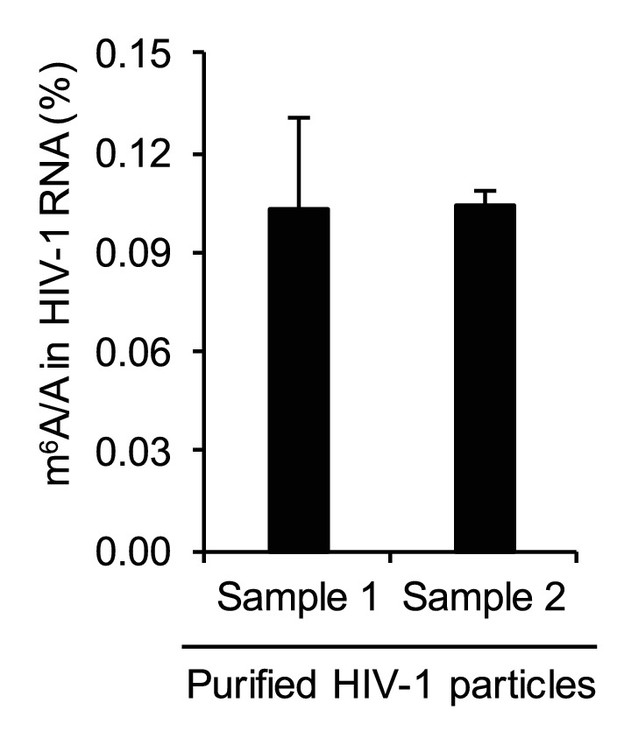
Quantification of HIV-1 RNA m6A level using liquid chromatography-mass spectrometry.
HIV-1 RNA (250 ng) was isolated from highly purified HIV-1MN virions (total 600 μg of p24 capsid) and subjected to quantitative analysis of the m6A level using LC-MS/MS (n=3 of each sample). The results are presented are from representative of two independent experiments.
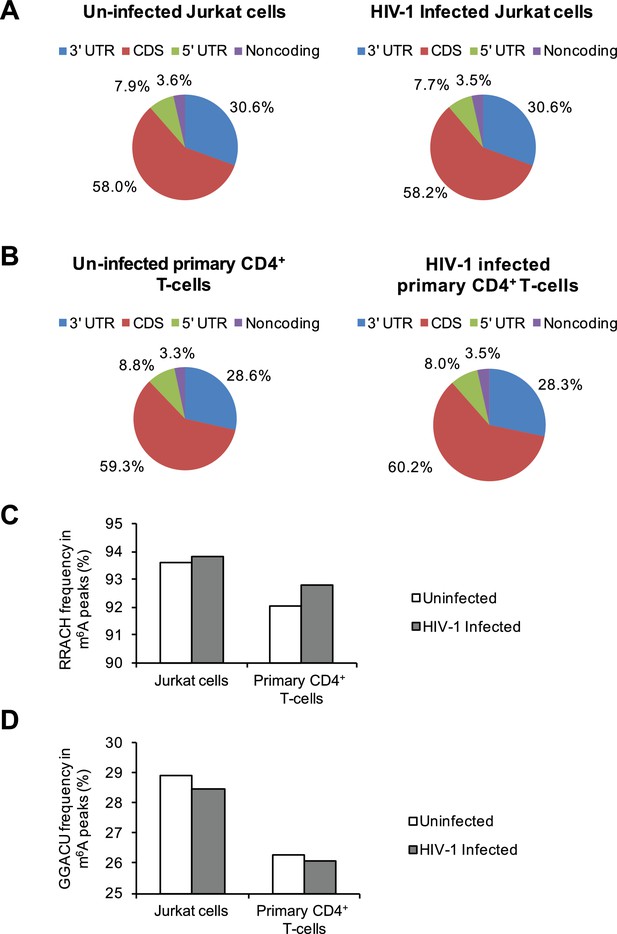
Distribution of m6A in cellular RNAs and the frequency of m6A motifs in HIV-1-infected cells.
(A–B) Pie charts show the distribution of m6A peaks in the 5′ UTR, coding DNA sequence (CDS), 3′ UTR, and noncoding regions of transcripts from uninfected and HIV-1-infected Jurkat T-cells (A) or primary CD4+ T-cells (B). The m6A peak distribution in HIV-1-specific RNAs is also shown. (C–D) Frequency of the RRACH motif (C) and the GGACU motif (D) within the m6A peaks in cellular RNAs from the uninfected control and HIV-1-infected cells. Data presented are the average results of duplicated samples (n=2).
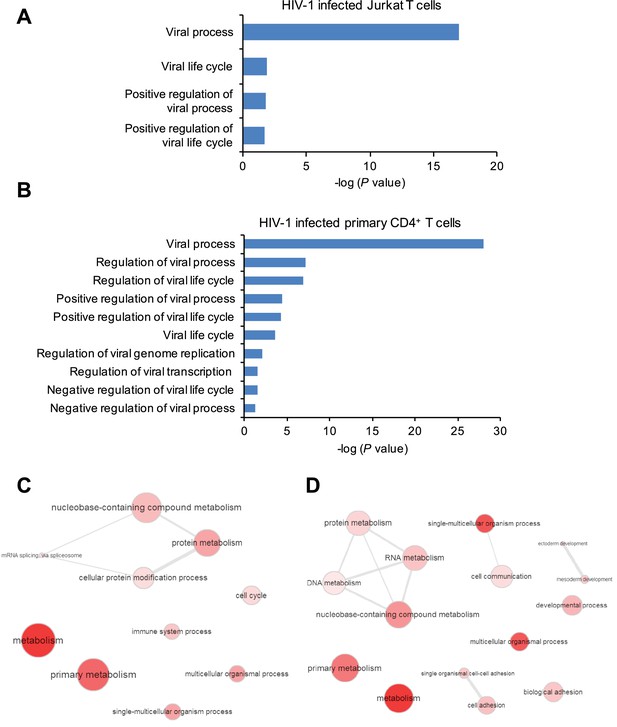
Gene ontology (GO) analysis of m6A-modified cellular genes in HIV-1 infected cells.
(A and B) GO terms specific to virus related pathways and corresponding p values, clustered from methylated genes detected in Jurkat cells (A) or primary CD4+ T cells (B) infected with HIV-1. (C and D) GO graphs showing functional clusters from genes with unique m6A peaks identified in HIV-1-infected Jurkat cells (C) or primary CD4+ T-cells (D) when compared to uninfected cells. Data presented are the average results of duplicated samples (n=2).
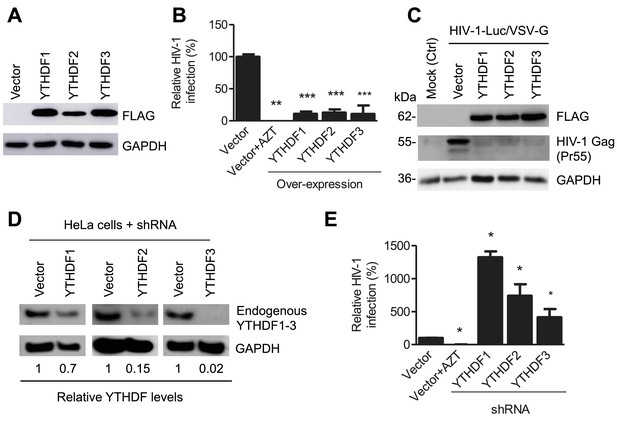
YTHDF1–3 proteins negatively regulate post-entry HIV-1 infection in HeLa cells.
(A–B) Overexpression of YTHDF1–3 proteins in HeLa cells significantly inhibits HIV-1 infection compared to vector control cells. (A) Overexpression of YTHDF1–3 proteins in HeLa cells was confirmed by immunoblotting. (B) HeLa cells overexpressing YTHDF1–3 proteins were infected with HIV-1 Luc/VSV-G at an MOI of 0.5 and viral infection was measured by luciferase activity at 24 hpi. (C) Overexpression of YTHDF1–3 proteins inhibits HIV-1 Gag protein synthesis in infected cells. HeLa cells overexpressing individual YTHDF1–3 proteins or the vector control cells were infected by HIV-1-Luc/VSV-G at an MOI of 0.5. At 24 hpi, the expression of HIV-1 Gag and YTHDF1–3 proteins (FLAG-tagged) was determined using immunoblotting. GAPDH was used as a loading control and mock-infected vector control cells were used as a negative control. (D and E) Individual knockdown of endogenous YTHDF1–3 proteins in HeLa cells significantly increases HIV-1 infection compared to vector control cells. HIV-1 infection assays were performed as described for panel B. *p<0.05, **p<0.005, and ***p<0.0005, compared to vector control without AZT treatment. All results are shown as mean ± SD (n=3) and data presented are representative of at least three independent experiments.
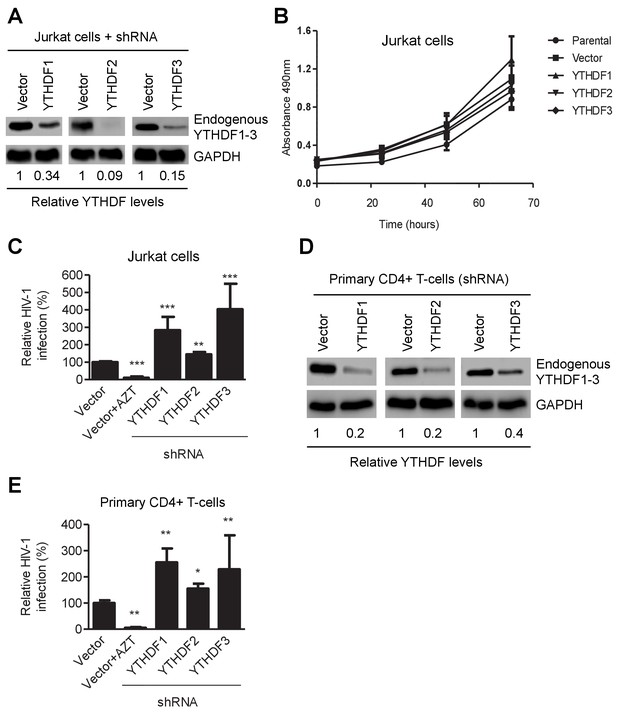
YTHDF1–3 proteins negatively regulate post-entry HIV-1 infection in CD4+ T-cells.
(A) Individual knockdown of endogenous YTHDF1–3 proteins in Jurkat CD4+ T cells was confirmed by immunoblotting. (B) Knockdown of YTHDF1–3 proteins does not affect proliferation of Jurkat cells. Jurkat cells (2 × 104) were seeded and cultured for 3 days. At the times indicated, cell proliferation was measured using the MTS assay. (C) Knockdown of YTHDF1–3 proteins significantly increases HIV-1 infection compared to vector control cells. (D) Individual knockdown of YTHDF1–3 proteins in activated primary CD4+ T-cells from a healthy donor. (E) Knockdown of YTHDF1–3 proteins significantly increases HIV-1 infection compared to vector control cells. (A and D) GAPDH was used as a loading control. (C and E) The vector controls without AZT were set as 100%. The reverse transcriptase inhibitor AZT treated cells were used as positive control for productive HIV-1 infection. *p<0.05, **p<0.005, and ***p<0.0005, compared to vector control without AZT treatment. All results are shown as mean ± SD (n=3) and data presented are representative of at least three independent experiments.
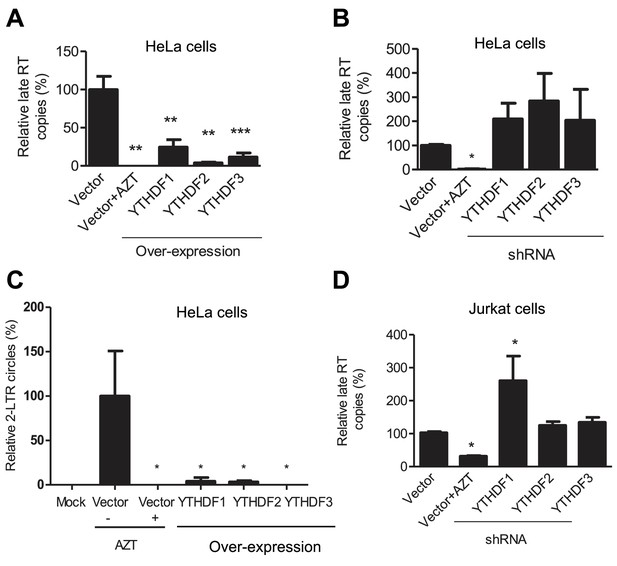
YTHDF1–3 proteins inhibit post-entry HIV-1 infection by blocking viral reverse transcription.
HeLa cells over-expressing or knocking-down (shRNA) individual YTHDF1–3 proteins were infected with HIV-1-Luc/VSV-G at an MOI of 0.5. (A, B and D) Genomic DNA was isolated from the cells 24 hr post-infection and HIV-1 late reverse transcription (RT) products were quantified by qPCR. (C) YTHDF family proteins reduce the formation of HIV-1 2-LTR circles in infected HeLa cells. At 24 hr post-infection, DNA was isolated from the cells and the 2-LTR circles were analyzed by qPCR and normalized to GAPDH levels. AZT treated vector control cells were used as a negative control for HIV-1 inhibition. *p<0.05 compared to the vector control without AZT treatment. All results are shown as mean ± SD (n=3) and data presented are representative of at least three independent experiments.
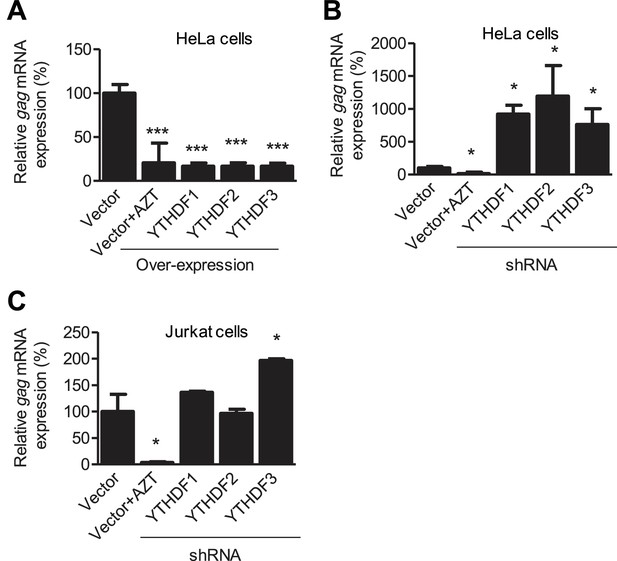
YTHDF1–3 proteins negatively regulate HIV-1 gag mRNA expression.
Specific shRNAs or scrambled shRNA vector-treated cells were infected with HIV-1 Luc/VSV-G at an MOI of 0.5. Total RNA was isolated from the cells 24 hr post-infection and HIV-1 gag mRNA levels were quantified using qRT-PCR. (A and B) HIV-1 gag mRNA levels in the infected HeLa cells with overexpression (A) or knockdown (B, shRNA) of YTHDF1–3 proteins. (C) HIV-1 gag mRNA levels in the HIV-1 infected Jurkat cells after knockdown of YTHDF1–3 proteins. AZT treated vector control cells were used as a negative control of HIV-1 infection (A–C). The vector controls without AZT were set as 100%. *p<0.05, **p<0.005, and ***p<0.0005, compared to vector control without AZT treatment. All results are shown as mean ± SD (n=3) and data presented are representative of at least three independent experiments.
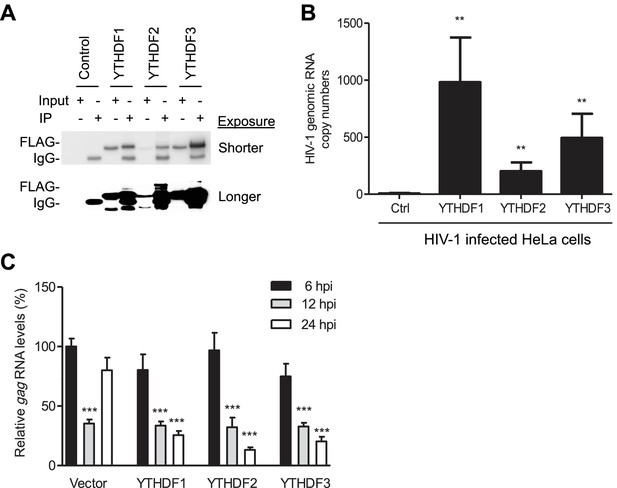
YTHDF1–3 proteins bind to HIV-1 gRNA in infected cells.
(A) Immunoblotting of YTHDF1–3 proteins in the input and immunoprecipitation (IP) samples from HIV-1-Luc/VSV-G infected HeLa cells. FLAG antibodies were used to immunoprecipitate FLAG-tagged YTHDF1–3 proteins overexpressed in HeLa cells after HIV-1 infection. A short and long exposure of the immunoblot is shown. (B) HIV-1 gRNA is bound by YTHDF1–3 proteins expressed in HeLa cells. HeLa cells stably overexpressing FLAG-tagged YTHDF1–3 proteins or empty vector control cells (Ctrl) were infected with HIV-1-Luc/VSV-G at an MOI of 5 for 3 hr. Cell lysates were immunoprecipitated with anti-FLAG, RNA was extracted and HIV-1 gag RNA levels were measured. **p<0.005 compared to the vector control cells. (C) YTHDF1–3 affect HIV-1 gag RNA kinetics. HIV-1 gag RNA levels in YTHDF1–3-expressing HeLa cells were quantified by qRT-PCR. The relative levels of gag RNA in infected cells were normalized to that of the vector control cells at 6 hr post-infection (hpi). ***p<0.0005, compared to the control cells at 6 hpi (set as 100%). All results are shown as mean ± SD (n=3) and data presented are representative of at least three independent experiments.
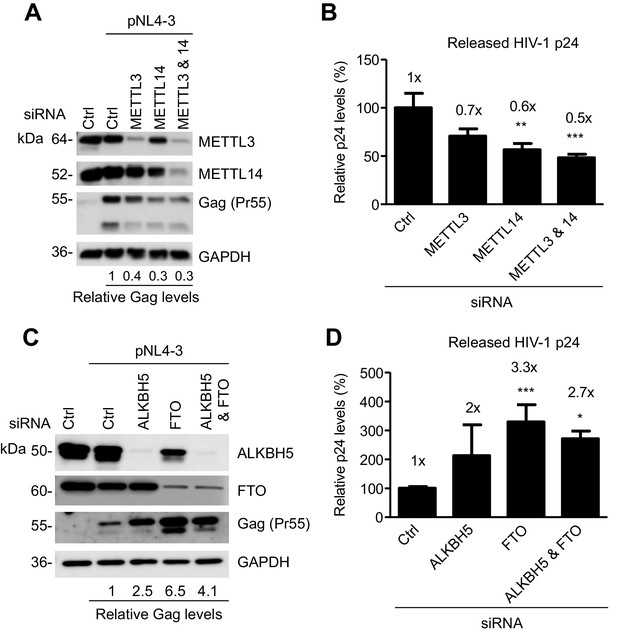
The m6A writers and erasers affect HIV-1 Gag expression in virus-producing cells.
(A and B) Individual or combined knockdown of endogenous METTL3 and METTL14 inhibits HIV-1 Gag protein expression. HEK293T cells were transfected with indicated siRNA, and then with an HIV-1 proviral DNA plasmid (pNL4-3). Cells and supernatants were collected for analyses at 36 hr post-transfection. (A) Expression of METTL3, METTL14 and HIV-1 Gag proteins in the transfected HEK293T cells was detected by immunoblotting. (C and D) Knockdown of endogenous AlkBH5, FTO, or both promotes HIV-1 Gag protein expression. HEK293T cells were transfected with indicated siRNA, and then with pNL4-3. Cells and supernatants were collected at 36 hr post-transfection. (C) Expression of AlkBH5, FTO and HIV-1 Gag proteins in the cells was detected by immunoblotting. (A and C) GAPDH was used as a loading control. Relative levels of Gag expression were normalized to GAPDH levels. (B and D) HIV-1 capsid p24 levels in supernatants were measured by ELISA. The relative levels (%) are also shown. *p<0.05 compared to the siRNA control. The results are shown as mean ± SD (n=3) and data presented are representative of three independent experiments.
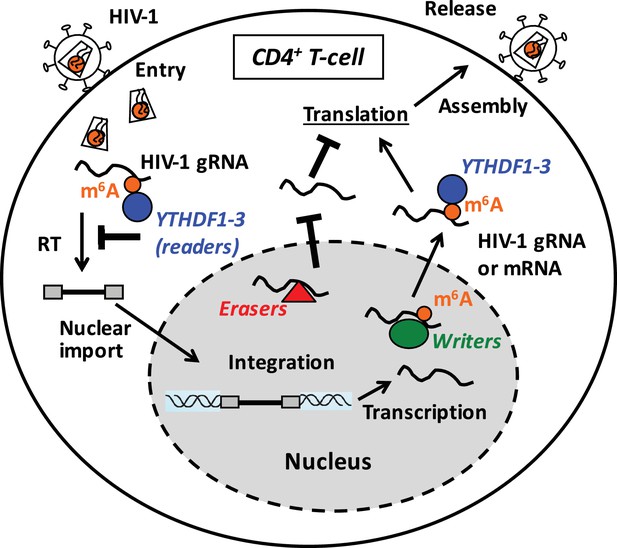
Proposed mechanisms and dynamics of m6A modification of HIV-1 RNA in regulating viral infection in cells.
In the nucleus, the m6A writers (METTL3 and METTL14) add the m6A marker to HIV-1 genomic RNA (gRNA) or mRNA, and the m6A erasers (FTO and AlkBH5) remove the m6A modifications of HIV-1 RNA. The m6A modification of HIV-1 RNA can promote viral protein translation in cells. In contrast, cytoplasmic m6A readers (YTHDF1–3) bind to m6A-modified HIV-1 gRNA, which can result in inhibition of HIV-1 reverse transcription (RT), viral mRNA expression, and thereby HIV-1 infection in cells.
Tables
The shRNA sequences used in this study.
| shRNA | Sequences (5’-3’) |
|---|---|
| Non-specific (vector) control | CCGGCAACAAGATGAAGAGCACCAACTCGAGTTGGTGCTCTTCATCTTGTTGTTTTT |
| YTHDF1 | CCGGCCCGAAAGAGTTTGAGTGGAACTCGAGTTCCACTCAAACTCTTTCGGGTTTTTG |
| YTHDF2 | CCGGCGGTCCATTAATAACTATAACCTCGAGGTTATAGTTATTAATGGACCGTTTTTG |
| YTHDF3 | CCGGGATAAGTGGAAGGGCAAATTTCTCGAGAAATTTGCCCTTCCACTTATCTTTTTG |
The siRNA sequences used in this study.
| siRNA | Sequences (5’-3’) |
|---|---|
| METTL3 | 5’-CTGCAAGTATGTTCACTATGA-3’ 5’-AGGAGCCAGCCAAGAAATCAA-3’ |
| METTL14 | 5’-TGGTGCCGTGTTAAATAGCAA-3’ 5’-AAGGATGAGTTAATAGCTAAA-3’ |
| FTO | 5’-AAATAGCCGCTGCTTGTGAGA-3’ |
| AlkBH5 | 5’-AAACAAGTACTTCTTCGGCGA-3’ |
Sequences of PCR primers and probes used in this study.
| Primers | Sequences (5’-3’) |
|---|---|
| HIV-1 gag forward | CTAGAACGATTCGCAGTTAATCCT |
| HIV-1 gag reverse | CTATCCTTTGATGCACACAATAGAG |
| Unspliced GAPDH forward | GGGAAGCTCAAGGGAGATAAAATTC |
| Unspliced GAPDH reverse | GTAGTTGAGGTCAATGAAGGGGTC |
| Spliced GAPDH forward | GGAAGGTGAAGGTCGGAGTCAACGG |
| Spliced GAPDH reverse | CTGTTGTCATACTTCTCATGGTTCAC |
| MH531 forward (for HIV-1 late reverse transcription (RT) products) | TGTGTGCCCGTCTGTTGTGT |
| BB reverse (for late RT products) | GGATTAACTGCGAATCGTTC |
| HIV-1 late RT product probe | TCGACGCAGGACTCGGCTTGCT |
| 2-LTR probe | AAGTAGTGTGTGCCCGTCTGTTGTGTGACTC |
| 2-LTR forward | GCCTGGGAGCTCTCTGGCTAA |
| 2-LTR reverse | GCCTTGTGTGTGGTAGATCCA |
| LW59 (forward, alternative for late RT detection in shRNA vector-transduced cells) | GACATAGCAGGAACTACTAGTACCC |
| LW60 (reverse, alternative for late RT detection in shRNA vector-transduced cells) | GGTCCTTGTCTTATGTCCAGAATGC |





