Beneficial impacts of neuromuscular electrical stimulation on muscle structure and function in the zebrafish model of Duchenne muscular dystrophy
Figures
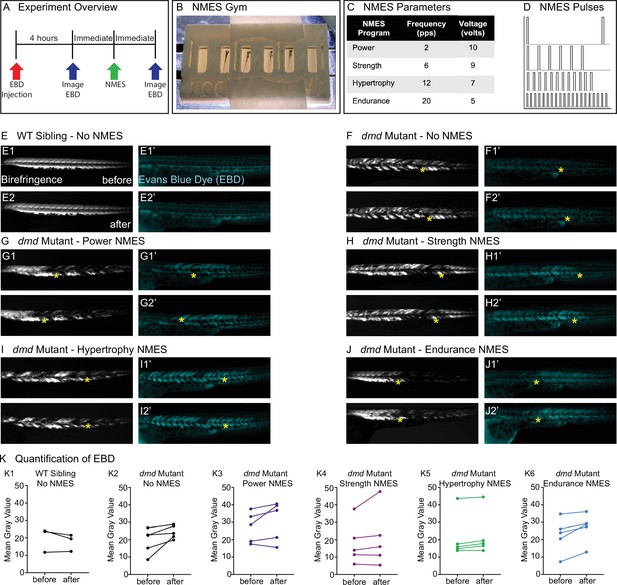
Four neuromuscular electrical stimulation (NMES) paradigms do not result in immediate damage to the sarcolemma.
(A) Experimental overview. At 2 days post-fertilization (dpf), WT siblings and dmd mutants were injected with Evans blue dye (EBD). 4 hr later, zebrafish were imaged for birefringence and EBD before and after a single session of NMES. (B) For NMES, zebrafish are placed in a 3D-printed gym with their heads towards the positive electrode and tails towards the negative electrode. (C, D) NMES delivers a series of square wave pulses that vary in frequency and voltage. We named these paradigms after weightlifting regimes. (E–J) Anterior left, dorsal top, side-mounted birefringence, and EBD fluorescent images. Yellow asterisks denote the same position in embryos before and after NMES. (E) WT sibling control exhibits healthy muscle segments (E1, E2) and no dye entry in the muscle (E1’, E2’) during the first and second imaging sessions. (F) dmd mutant control has significant areas of degenerated muscle (F1) and dye entry (F1’) but no new areas of degeneration or dye entry during the second imaging session (F2, F2’). (G–J) Similar to the dmd mutant control, dmd mutants that receive NMES have significant areas of degenerated muscle and dye entry prior to NMES but no new areas of degeneration or dye entry during following NMES. (K) Quantification of EBD during the first and second imaging sessions.
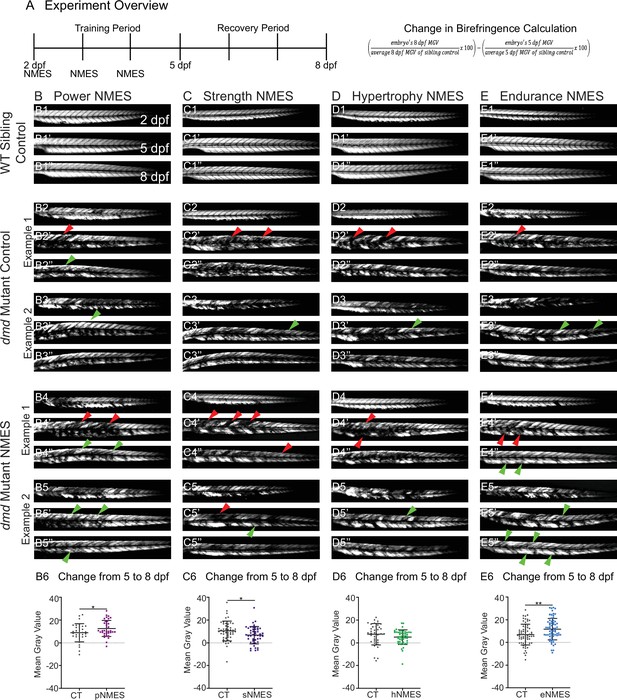
Impacts of neuromuscular electrical stimulation (NMES) paradigms on muscle structure through time.
(A) Experimental overview and calculation of change in mean gray value from 5 to 8 days post-fertilization (dpf). At 2 dpf, birefringence images were taken followed by the first session of NMES. At 3 and 4 dpf, zebrafish underwent the second and third sessions of NMES, respectively. Birefringence images were taken at 5 and 8 dpf. The training program was divided into the training period (2–4 dpf) and the recovery period (5–8 dpf). (B–E) Anterior left, dorsal top, side-mounted birefringence images for WT sibling controls (panels labeled 1), control dmd mutants (two examples shown, panels labeled 2 and 3), and NMES-treated dmd mutants (two examples shown, panels labeled 4 and 5). The NMES regimens are labeled as such: panels labeled B were treated with power NMES (pNMES), panels labeled C were treated with strength NMES (sNMES), panels labeled D were treated with hypertrophy NMES (hNMES), and panels labeled E were treated with endurance NMES (eNMES). The change in mean gray values from 5 dpf to 8 dpf represents how the muscle responds to and recovers from three sessions of NMES and is shown in panels labeled 6. Positive changes indicate improvements in muscle structure while negative changes indicate deterioration in muscle structure. Red arrowheads denote degeneration from the previous time point, green arrowheads denote regeneration from the previous time point. pNMES (B6, maroon squares) and eNMES (E6, blue squares) significantly improve muscle structure in dmd mutants compared to dmd mutant controls (gray circles). sNMES (C6, purple squares) significantly worsens muscle structure in dmd mutants while hNMES (D6, green squares) trends to decrease muscle structure compared to dmd mutant controls (gray circles). Each data point represents a single zebrafish. Birefringence data were analyzed using two-sided t-tests. *p<0.05, **p<0.01.
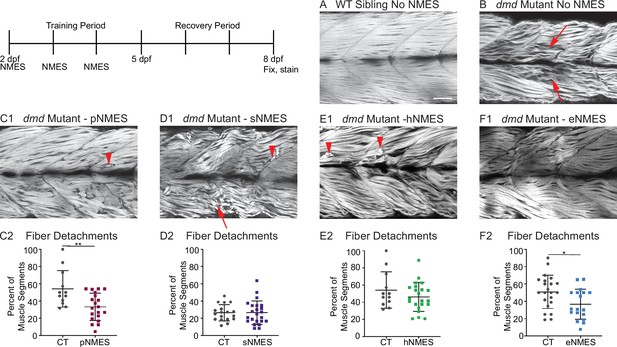
Impacts of neuromuscular electrical stimulation (NMES) paradigms on muscle fiber structure and degeneration.
Phalloidin staining for F-actin at 8 days post-fertilization (dpf) allows for visualization of individual muscle fibers and the ability to count detached fibers in dmd mutants. Anterior left, dorsal top, side mounted. Red arrows point to disorganized muscle fibers, and red arrowheads point to detached muscle fibers. (A) Representative image of WT sibling demonstrates organized muscle fibers with well-defined myotome boundaries. (B) Representative image of dmd mutants demonstrates disorganized, wavy muscle fibers with poorly defined myotome boundaries and empty space between individual muscle fibers. (C1) Representative image of dmd mutant that received power NMES (pNMES) demonstrates less muscle fiber waviness, lack of empty space between muscle fibers but visible detached fibers. (D1) Representative image of dmd mutant that received strength NMES (sNMES) demonstrates massive deterioration of muscle fiber structure, disorganized myotomes with poorly defined boundaries. (E1) Representative image of dmd mutant that received hypertrophy NMES (hNMES) demonstrates improved muscle fiber organization with more defined myotome boundaries but visibly detached muscle fibers and empty space between fibers. (F1) Representative image of dmd mutant that received endurance NMES (eNMES) demonstrates healthy myotomes with clearly defined boundaries, organized muscle fibers with very few wavy fibers, and lack of empty space between fibers. Quantification of the percentage of muscle segments with detachments indicates that pNMES (C2) and eNMES (F2) significantly reduce fiber detachments in dmd mutants. sNMES (D2) and hNMES (E2) do not impact the percent of muscle segments with detachments. Each data point represents a single fish. A muscle segment is defined as half of a myotome. Muscle detachment data were analyzed using two-sided t-tests. *p<0.05, **p<0.01. Scale bar is 50 µm.
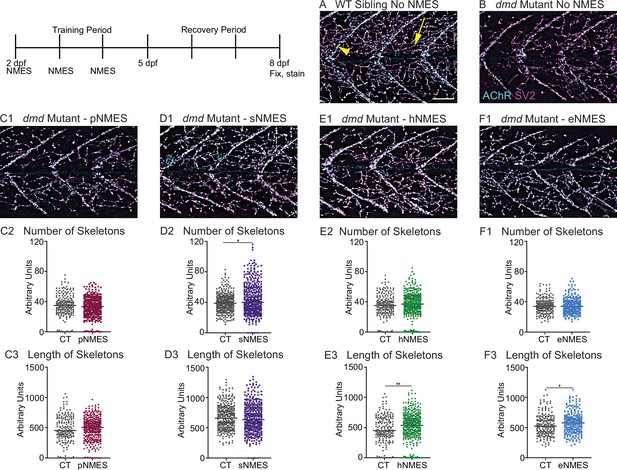
Impacts of neuromuscular electrical stimulation (NMES) paradigms on neuromuscular junction (NMJ) structure.
Anti-SV2 (cyan) alpha-bungarotoxin (AChR; magenta) visualize the pre- and postsynaptic components of the NMJ, respectively. (A) Representative image of WT sibling. Myoseptal innervation, innervation at the chevron-shaped myotendinous junction (yellow arrowhead) is slow-twitch innervation. Fast-twitch muscle innervation is the network between the MTJs, yellow arrow points to fast-twitch muscle innervation. (B) Representative image of dmd mutant demonstrates a visible reduction in innervation, with relatively large portions of the muscle segments lacking innervation. (C1, D1, E1, F6) Representative images of dmd mutants that completed three sessions of the NMES paradigms. NMJ images were skeletonized as previously described. Strength NMES(sNMES) was the only paradigm that increased the number of skeletons (D2). Both hypertrophy NMES (hNMES) and endurance NMES (eNMES) increase skeleton length (E3, F3). Power NMES did not change the number or length of skeletons compared to dmd mutant controls (C2,3). Scale bar is 50 µm. NMJ data were analyzed using either an ordinary one-way ANOVA with Tukey’s multiple comparisons test or a Kruskal–Wallis test with Dunn’s multiple-comparison test. **p<0.01, ***p<0.001, ****p<0.0001.
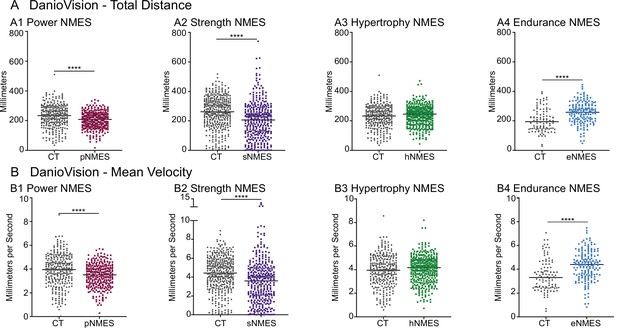
Impacts of neuromuscular electrical stimulation (NMES) paradigms on swimming.
DanioVision was used to assess the impact of NMES on total distance (A) and (B) mean velocity. Measurements were made at 8 days post-fertilization (dpf). (A1, B1) dmd mutants that completed power NMES (pNMES) exhibited significant reductions in total distance and mean velocity compared to dmd mutants in the control group. (A2, B2) Strength NMES also negatively affected swimming activity in dmd mutants compared to control dmd mutants. (A3, B3) No change in total distance or mean velocity is observed following hypertrophy NMES (hNMES). (A4, B4) dmd mutants that completed endurance NMES (eNMES) swam a significantly greater total distance and at a significantly faster mean velocity compared to dmd mutants in the control group. Each data point represents a single time point for an individual zebrafish. Each zebrafish has a total of 15 points. DanioVision data were analyzed using two-sided t-tests. **p<0.01, ***p<0.001, ****p<0.0001.
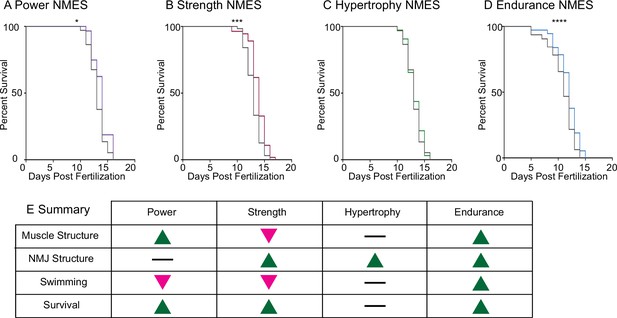
Impacts of neuromuscular electrical stimulation (NMES) paradigms on survival.
Survival was tracked following completion of the three NMES sessions. Survival was significantly improved in dmd mutants that completed power (A), strength (B), and endurance (C) NMES. (D) Hypertrophy NMES had no effect on survival in dmd mutants. Survival data were analyzed using a Mantel–Cox test. *p<0.05, ***p<0.001, ****p<0.0001. (E) Summary of the impacts of NMES paradigms on neuromuscular structure and function. Note that endurance NMES is the only paradigm that improved all aspects.
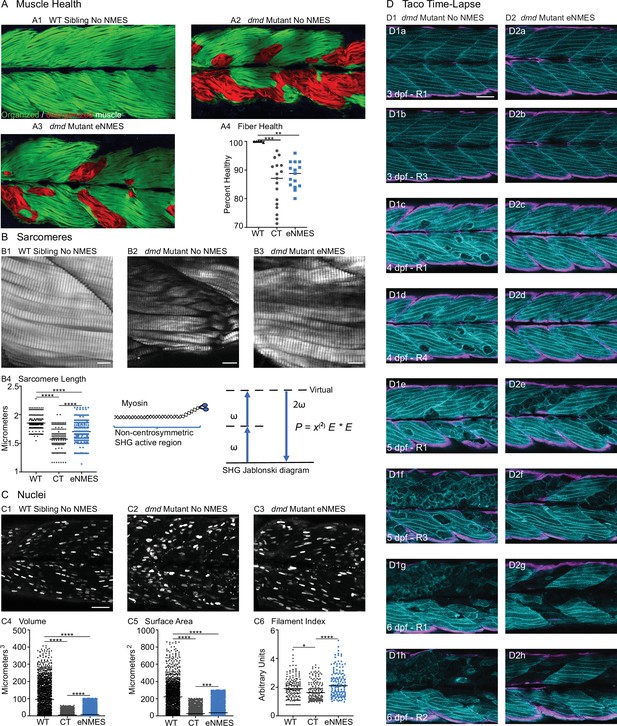
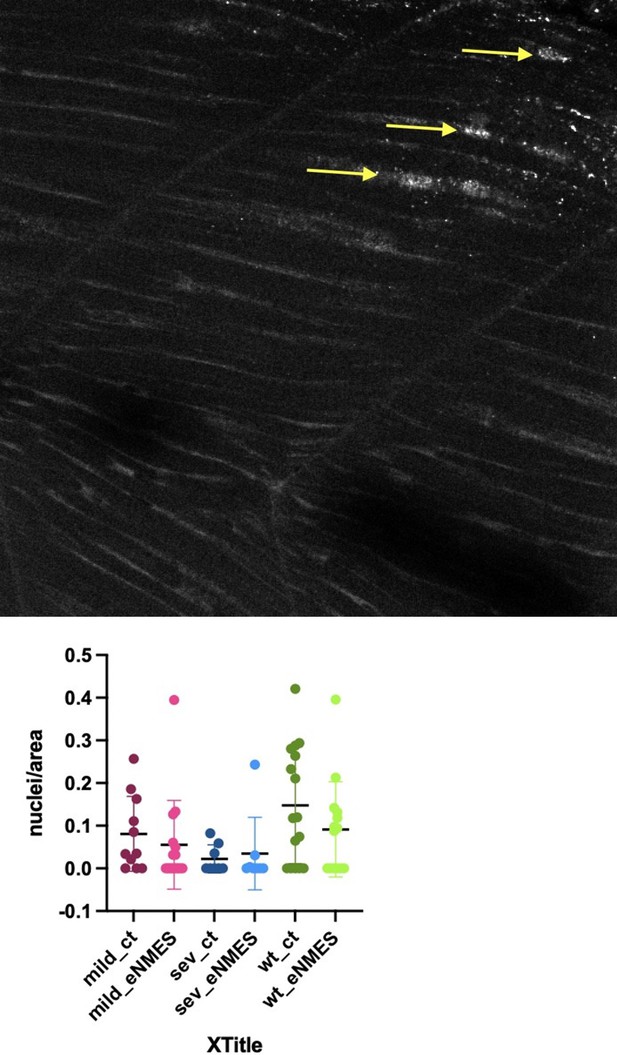
Endurance neuromuscular electrical stimulation (eNMES) improves multiple components of muscle health.
(A) Machine learning was used to quantify muscle health pixel-by-pixel. Green indicates healthy pixels while red indicates unhealthy pixels. (B) Second harmonic generation (SHG) microscopy was used to quantify sarcomere length at 8 days post-fertilization (dpf). Representative SHG images of WT sibling control (B1), dmd mutant controls (B2), and dmd mutants that completed eNMES training. Anterior left, dorsal top, side mounted. Scale bars are 10 µm. (B4) Sarcomere length is significantly shorter in dmd mutant controls compared to WT sibling controls. However, eNMES significantly improves sarcomere length, bringing it closer to WT lengths. Each point represents a single sarcomere along a predetermined length of a muscle fiber. Multiple muscle fibers were measured per zebrafish. (C) Muscle nuclei were imaged at 8 dpf as a potential mechanism for improved muscle health. Anterior left, dorsal top, side mounted. (C1) Representative image of WT sibling control demonstrates healthy ellipsoidal nuclei organized along the length of the muscle fibers. (C2) Representative image of dmd mutant control demonstrates fragmented punctae as well as more spherical nuclei that clustering within the muscle segments. (C3) Representative image of dmd mutant that completed eNMES training demonstrates healthier, ellipsoidal nuclei that appear more organized within the muscle segments. Quantification of nuclear size indicates that eNMES significantly increases the volume (C4) and surface area (C5) of muscle nuclei compared to dmd mutant controls. However, nuclei are still significantly smaller compared to WT sibling controls, visually appearing to have an increased number of myonuclei compared to unstimulated dmd mutants. (C6) Filament index was used to assess circularity, specifically the departure from a circle. Filament index is significantly higher in dmd mutants that completed eNMES training, indicating that nuclei are more elongated compared to dmd mutant controls. Each point represents a single nuclei within a z-stack. (D) Transgenic dmd mutants (mylpfa:lyn-cyan [cyan], smych1:GFP [magenta]) were used to visualize changes in structural integrity of fast- and slow-twitch muscle fibers across three days. Anterior left, dorsal top, side mounted. Scale bar is 50 µm. Images were taken around the 12th myotome. (D1) Representative dmd mutant control. (D1a–D1b). At 3 dpf, there is no dystrophy in the imaged myotomes. (D1c–D1e) At 4 dpf and the beginning of 5 dpf, dystrophy is minimal with relatively few detaching muscle fibers. (D1f) However, massive muscle degeneration occurs between the first found of imaging and the third round of imaging at 5 dpf. (D1g–D1h) Fiber degeneration is still present, suggesting that the damaged muscle fibers have not been cleared and regeneration is unlikely. (D2) Representative dmd mutant that is undergoing eNMES training. (D2a–D2b) The first session of eNMES at 3 dpf does not result in immediate damage to the muscle. (D2c–D2d) Similarly, following the second session of eNMES at 4 dpf, there is no immediate muscle damage occurring in the imaged myotomes. (D2e) At 5 dpf, following the third session of eNMES, muscle fiber degeneration is evident, but by the third round of imaging (D2f), these damaged areas are being cleared and there is evidence of regeneration. (D2g–D2h) At 6 dpf, previously damaged muscle segments have new muscle fibers present. All data were analyzed using either an ordinary one-way ANOVA with Tukey’s multiple comparisons test or a Kruskal–Wallis test with Dunn’s multiple-comparison test. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
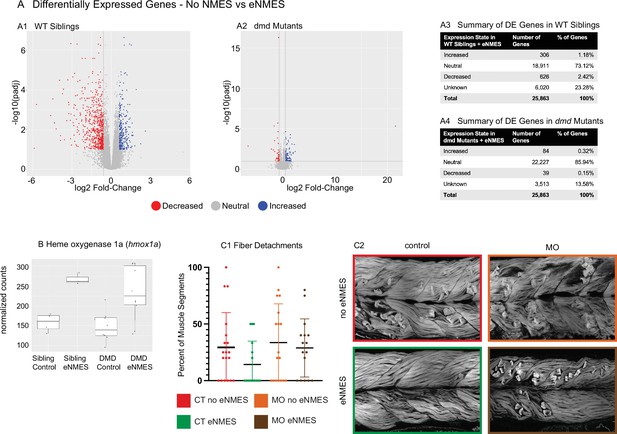
dmd mutants do not respond to endurance neuromuscular electrical stimulation (eNMES) in the same manner as WT siblings and heme oxygenase is required for eNMES-mediated improvement.
RNAseq analysis was performed at 7 days post-fertilization (dpf) in WT siblings and dmd mutants that completed eNMES training, and their expression patterns were compared with their respective controls. (A1) Volcano plot showing significantly and biologically upregulated (blue dots) and downregulated (red dots) genes in WT siblings that completed eNMES versus those that did not. (A2) Volcano plot showing significantly and biologically upregulated (blue dots) and downregulated (red dogs) genes in dmd mutants that completed eNMES versus those that did not. (A3–A4) Summary of differentially expressed genes in WT siblings (A3) and dmd mutants (A4). WT siblings had 932 differentially expressed genes compared to 123 differentially expressed genes in dmd mutants, suggesting that dmd muscle responds differently to eNMES and the genes responsible for eliciting beneficial effects on muscle structure and function are different. (B) hmox1a expression was increased with eNMES in both WT and dmd mutants. (C1) Whereas eNMES significantly reduces the percentage of segments with dystrophy, dmd mutants injected with morpholinos against hmox1a do not show improvement with eNMES. (C2) Representative images of larvae showing that hmox1a is necessary for eNMES-mediated improvement of dmd muscle.
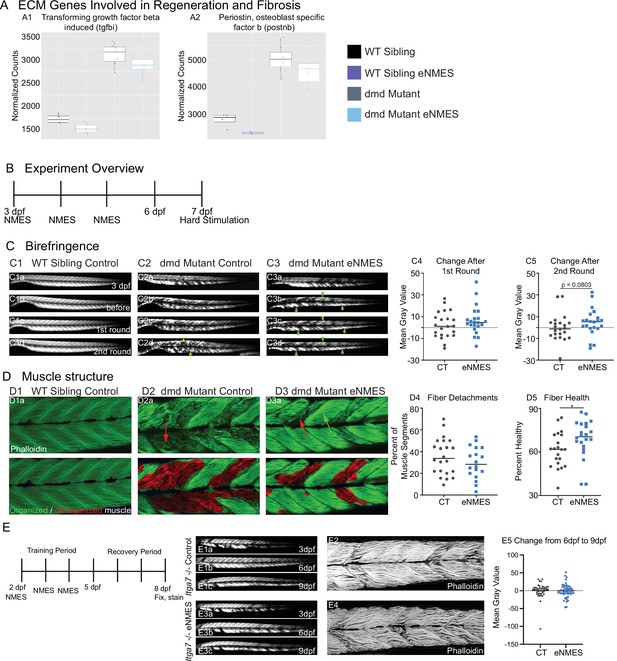
Muscle resilience to hard stimulation is increased with endurance neuromuscular electrical stimulation (eNMES), and Itga7 is required for eNMES-mediated improvement.
We identified three extracellular matrix (ECM) genes from RNAseq analysis, tgfbi (A1), postnb (A2), itgb1b.2 (not shown) that are significantly upregulated in dmd mutants compared to WT siblings and trend towards being downregulated with eNMES in dmd mutants. (B) Experimental overview. At 3 days post-fertilization (dpf) (disease onset), birefringence images were taken followed by the first session of eNMES. At 4 and 5 dpf, zebrafish undergo the second and third NMES sessions, respectively. At 7 dpf, muscle resilience was tested using a hard electrical stimulation paradigm intended to cause muscle damage. (C) Birefringence images were taken at 3 dpf (C1a). (C1b–d) Birefringence images were taken at 7 dpf before the first hard stimulation (C1b), after the first hard stimulation (C1c), and after the second hard stimulation (C1d). No visible changes in birefringence are observed in WT siblings after the two stimulation sessions. (C2) For dmd mutant controls, the first round of stimulation did not result in visible changes to birefringence (C2c), but, after the second round, areas of muscle degeneration are visible (C2d, yellow asterisks). Conversely, in dmd mutants that completed three sessions of eNMES, the first (C3c) and second (C3d) rounds of stimulation did not result in visible changes to birefringence (green arrowheads denote intact areas of birefringence that remain intact). (C4, C5) Change in birefringence from before to after the first round (C4) and second (C5) of stimulation suggests that eNMES training may improve muscle resilience. (D) Phalloidin was used to visualize individual muscle fibers. (D1a) Representative image of a WT sibling control demonstrates healthy, organized muscle fibers, and myotomes. (D2a) Representative image of a dmd mutant control highlights disorganized and wavy muscle fibers and fiber detachments. (D3a) Representative image of a dmd mutant that completed eNMES demonstrates some wavy muscle fibers and detached fibers intermixed with relatively healthy myotomes. (D4) The percent of muscle segments with detached fibers following the hard stimulation is reduced in dmd mutants that complete eNMES training compared to dmd mutant controls. For this analysis, a muscle segment was defined as half of a myotome. (D1b, D2b, D3b) Machine learning was used to quantify muscle health pixel-by-pixel. Green indicates healthy pixels while red indicates unhealthy pixels. (D5) The percent of healthy muscle following the hard stimulation is significantly higher in dmd mutants that completed eNMES compared to dmd mutant controls. (E) itga7 mutants were subjected to the same eNMES protocol that results in improvements in dmd mutants. Note that eNMES does not improve birefringence (panels E1, E3, quantified in E5) or muscle structure in itga7 mutants (E2, E4). All data were analyzed using two-sided t-tests. *p<0.05.
Summary.
Endurance neuromuscular electrical stimulation (eNMES) positively benefits neuromuscular health, function, and survival in dmd mutants. Muscle fibers in dmd mutants treated with eNMES are more organized and have fewer detachments. Neuromuscular junctions (NMJs) are longer in eNMES-treated dmd mutants. Sarcomeres are longer and nuclei are more ellipsoid and aligned. eNMES-treated mutants swim faster and more distance than control siblings. Both heme oxygenase and Itga7 are required for eNMES-mediated improvements.
Videos
Video of embryos undergoing different electrostimulation paradigms.
The paradigms are noted in the video.
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/62760/elife-62760-transrepform1-v1.docx
-
Source data 1
Source data are organized by figure with titles of the measurements in the columns and sheets are named with the appropriate figure.
Titles of panels also include the figure panel that the data is for.
- https://cdn.elifesciences.org/articles/62760/elife-62760-data1-v1.xlsx