The Spike D614G mutation increases SARS-CoV-2 infection of multiple human cell types
Figures
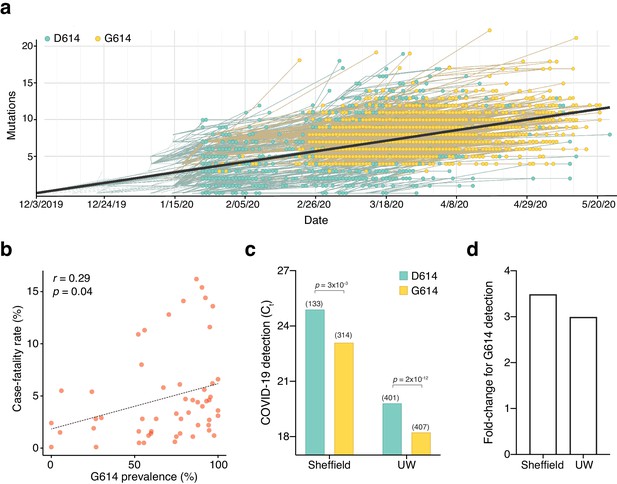
The SARS-CoV-2 D614G mutation has spread rapidly and is correlated with increased fatality and higher viral load.
(a) Prevalence of D614G-containing SARS-CoV-2 genomes over time. This visualization was produced by the Nextstrain webtool using GISAID genomes (n = 3,866 genomes samples from January 2020 to May 2020). (b) Per-country Pearson correlation of G614 prevalence with the case-fatality rate (n = 56 countries and 22,103 genomes). (c) Threshold cycle for quantitative polymerase chain reaction (qPCR) detection of SARS-CoV-2 from patients with D614 and G614 Spike. Numbers in parentheses indicate the number of COVID-19 patients in each group and significance testing is using the Wilcoxon rank sum test. This Sheffield data was originally presented in Korber et al., 2020. The University of Washington data was originally presented in Wagner et al., 2020. (d) Fold-change of increase in viral RNA present in COVID-19 patient samples with G614 Spike as compared to those with D614 Spike.
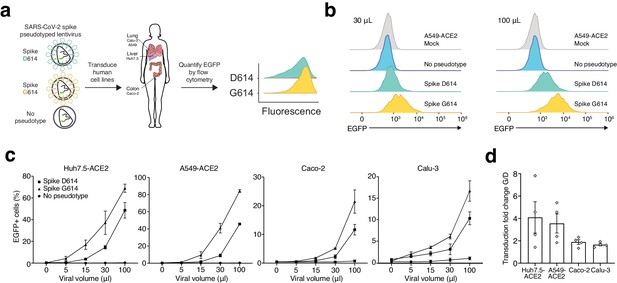
SARS-CoV-2 Spike D614G-pseudotyped lentivirus results in increased transduction of human lung, liver, and colon cell lines.
(a) Schematic of EGFP lentivirus pseudotyped with SARS-CoV-2 Spike proteins (or no pseudotype) and quantification of EGFP fluorescence by flow cytometry. (b) Flow cytometry of A549ACE2 cells at 3 days post-transduction with 30 or 100 µL SARS-CoV-2 Spike-pseudotyped lentivirus. (c) Percent of EGFP+ cells at 3 days post-transduction with the indicated volume of virus and pseudotype in human liver Huh7.5ACE2 cells, lung A549ACE2 cells, colon Caco-2 cells, and lung Calu-3 cells (n = 3 replicates, error bars are s.e.m.). (d) The fold-change for each viral transduction of G614 Spike over D614 Spike in four cell lines (error bars are s.e.m.).
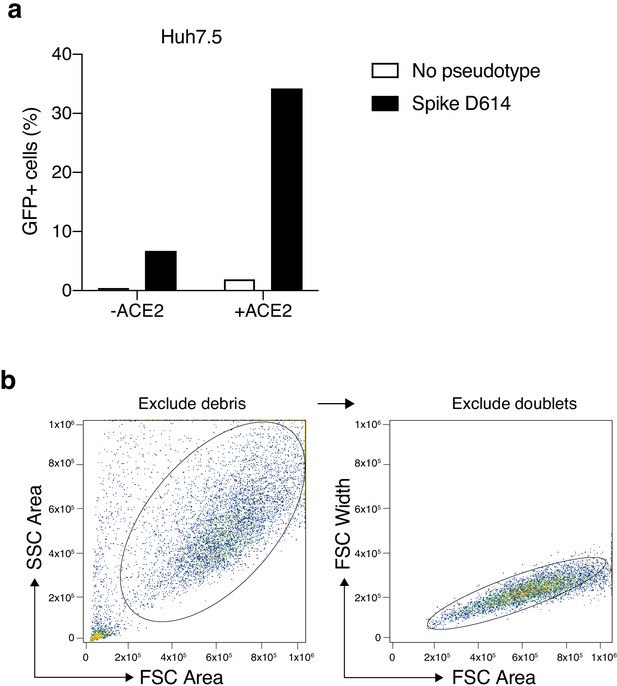
Increased transduction of SARS-CoV-2 Spike-pseudotyped lentivirus in human cells that constitutively overexpress the human ACE2 receptor.
(a) Percent of EGFP+ cells at 6 days post-transduction with 100 μL of supernatant SARS-CoV-2 Spike (D614) pseudotyped lentivirus and unpseudotyped lentivirus in human liver Huh7.5 with and without ACE2 overexpression. (b) Flow cytometry gating for GFP quantification. Cells were first gated by the forward and side scatter area to exclude debris and then doublets were excluded by gating on forward scatter area and width.
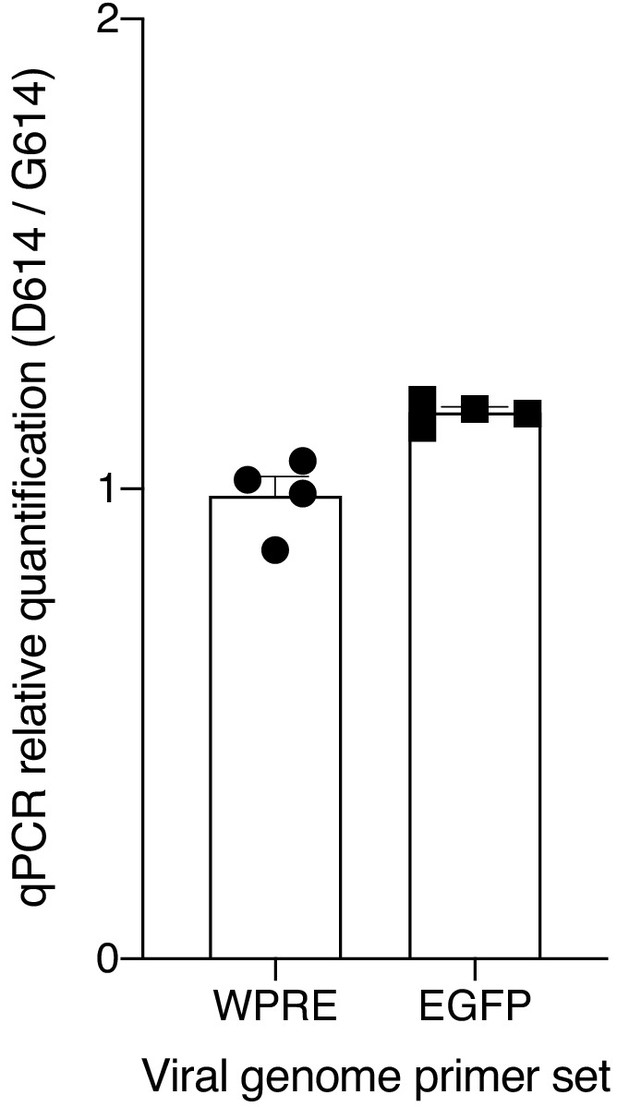
Quantitative PCR of viral RNA from SARS-CoV-2 Spike-pseudotyped lentiviruses.
Relative quantification (ΔΔCt) of viral RNA from SARS-CoV-2 Spike-pseudotyped lentiviruses. The primers amplify the woodchuck hepatitis virus posttranscriptional regulatory element (WPRE) or the EGFP gene in the viral RNA genome.
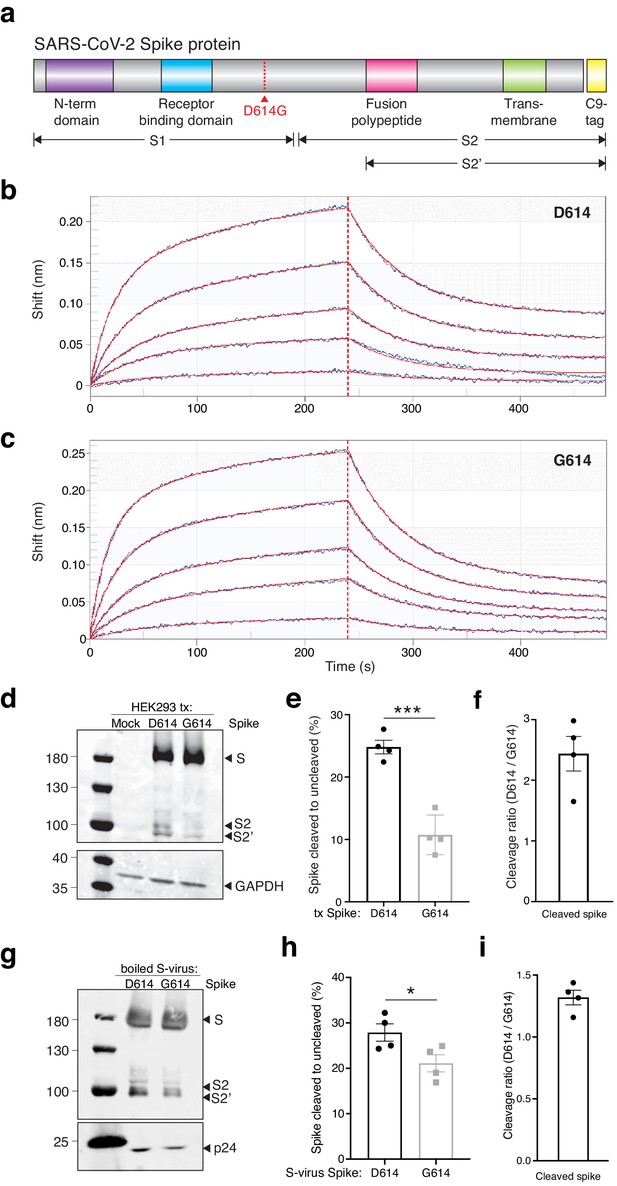
The SARS-CoV-2 Spike D614G variant displays similar ACE2 binding kinetics but altered proteolytic cleavage.
(a) Schematic diagram of SARS-CoV-2 Spike protein structure with the added C9 affinity tag on the C-terminus. Spike cleavage fragments S1, S2, and S2’ are also indicated. (b, c) Association and dissociation binding curves of Spike D614 (b) and Spike G614 (c) with hACE2. Blue curves represent Spike protein binding profiles at 200 nM, 100 nM, 50 nM, 25 nM, and 6.25 nM. Red curves represent the best global fit using a 2:1 heterogeneous ligand model. (d) Western blot of total protein lysate from HEK293FT cells after transfection with D614 Spike, G614 Spike, or mock transfection. (Upper) Detection of full-length Spike and cleavage fragments using an anti-C9 (rhodopsin) antibody. (Lower) Detection of GAPDH via anti-GAPDH antibody. (e) Fraction of cleaved (S2 + S2’) to uncleaved (full-length) fragments for Spike D614 and G614 (n = 4 replicates, error bars are s.e.m.). (f) Fold-change in cleavage between Spike variants (D614/G614) (n = 4 replicates, error bars are s.e.m.). (g) Western blot of boiled S-virus pseudotyped with D614 Spike or G614 Spike. (Upper) Detection of full-length Spike and cleavage fragments using an anti-C9 (rhodopsin) antibody. (Lower) Detection of p24 capsid protein via anti-24 antibody. (h) Fraction of cleaved (S2 + S2’) to uncleaved (full-length) fragments for Spike D614 and G614 on lentiviral particles (n = 4 replicates, error bars are s.e.m.). (i) Fold-change in cleavage between Spike variants on lentiviral particles (D614/G614) (n = 4 replicates, error bars are s.e.m.). *p≤0.05, ***p≤0.001. Significance testing was done with an unpaired two-tailed t-test.
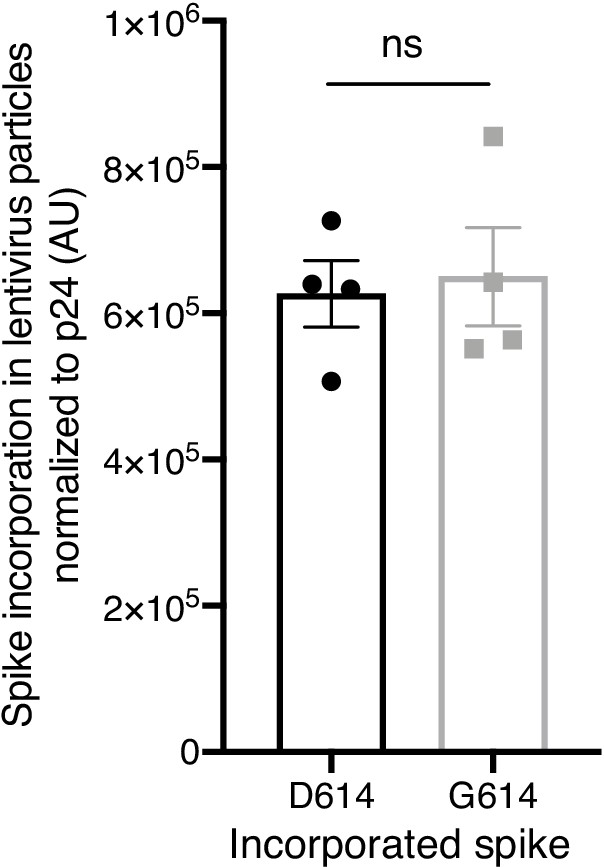
Spike incorporation into Spike-pseudotyped lentiviral particles.
Spike D614 and Spike G614 protein incorporation rate into pseudotyped lentiviral particles was quantified by densitometry of total Spike expression (cleaved and uncleaved) normalized to expression of the lentiviral capsid protein p24. Significance testing was done with an unpaired two-tailed t-test.
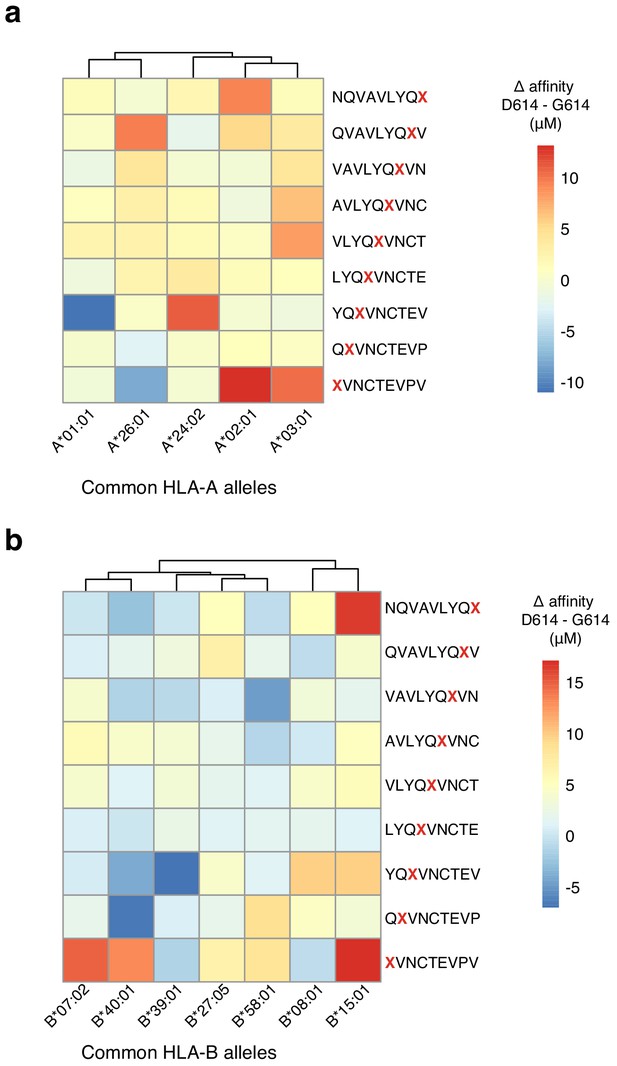
Change in MHC binding affinity for peptides near the D614G mutation in the SARS-CoV-2 Spike protein.
Predicted change in binding affinity for common HLA-A alleles (a) and common HLA-B alleles (b). Predictions were computed using the NetMHC software package.
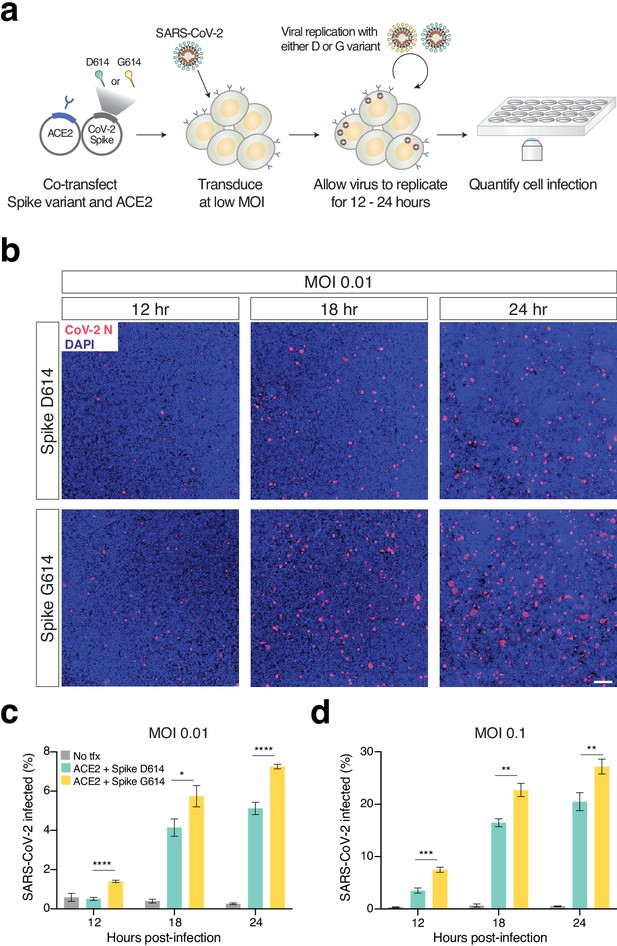
Increased infection of SARS-CoV-2 virus with the Spike D614G variant in human cells.
(a) Schematic diagram of trans-complementation assay to assess the impact of SARS-CoV-2 Spike variants in an isogenic fashion. Plasmids containing a single Spike variant and ACE2 are co-transfected into HEK293T cells and then, after 24 hr, are infected with SARS-CoV-2 (Isolate USA-WA1/2020). (b) Representative images of HEK293T cells infected with SARS-CoV-2 (multiplicity of infection [MOI]: 0.01) and fixed at the indicated time point post-infection. The cells were stained with DAPI (blue) and an antibody for SARS-CoV-2 nucleocapsid protein (red). (Top) Transfection of ACE2 and SARS-CoV-2 Spike D614 plasmids. (Bottom) Transfection of ACE2 and SARS-CoV-2 Spike G614 plasmids. Scale bar: 1 mm. (c, d) Percent of SARS-CoV-2-infected cells at 12, 18, or 24 hr post-infection as measured by immunocytochemistry for SARS-CoV-2 nucleocapsid (N) protein and quantification using an imaging cytometer. A MOI of 0.01 is in shown in (c) and a MOI of 0.1 is shown in (d). *p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001. Significance testing was done with an unpaired two-tailed t-test.
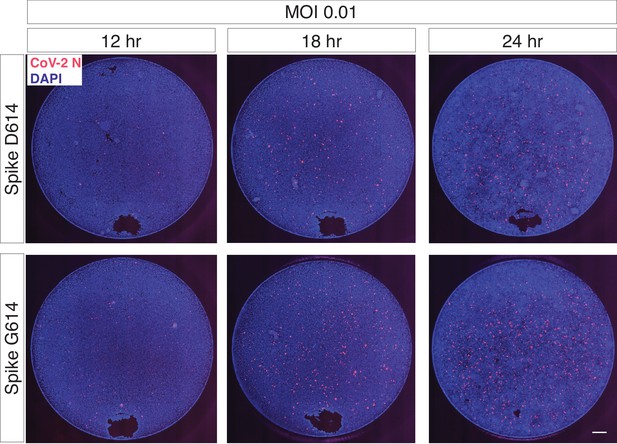
Trans-complementation of full SARS-CoV-2 virus with Spike G614 increases viral infection.
The regions shown in Figure 4b are representative sub-regions selected from the full images (entire well). The full images are shown here without any contrast adjustment for a single biological replicate (n = 6 replicate infections for each time point-MOI combination). DAPI (nuclei) are shown in blue, and SARS-CoV-2 nucleocapsid (N) protein is shown in red. Scale bar: 500 μm.
Tables
SARS-CoV-2 Spike D614 and G614 variants have comparable binding affinities for hACE2.
Values of kinetics assay from bio-layer interferometry with purified Spike and ACE2. A 2:1 model yields two KD values as measured by Kdis/Kon for each binding process.
| KD1 (nM) | Kon1 (1/M * s) | Kdis1 (1/s) | KD2 (nM) | Kon2 (1/M * s) | Kdis2 (1/s) | |
|---|---|---|---|---|---|---|
| Spike D614 | 8.45 | 3.51 × 104 | 2.96 × 10−4 | 127 | 1.59 × 105 | 2.01 × 10−2 |
| Spike G614 | 18.0 | 4.25 × 104 | 7.67 × 10−4 | 92.7 | 2.22 × 105 | 2.06 × 10−2 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (H. sapiens) | A549 | ATCC | Cat # CCL-185 RRID:CVCL_0023 | |
| Cell line (H. sapiens) | A549ACE2 | This study | N/A | |
| Cell line (H. sapiens) | HEK293FT | Thermo | Cat # R70007 RRID:CVCL_6911 | |
| Cell line (H. sapiens) | Huh-7.5 | tenOever lab | RRID:CVCL_7927 | |
| Cell line (H. sapiens) | Huh-7.5ACE2 | This study | N/A | |
| Cell line (H. sapiens) | Caco-2 | ATCC | Cat # HTB-37 RRID:CVCL_0025 | |
| Cell line (H. sapiens) | Calu-3 | tenOever Lab | Cat # HTB-55 RRID:CVCL_YZ47 | |
| Antibody | Anti-GAPDH (rabbit monoclonal) | Cell Signalling | Cat # 2118 RRID: AB_561053 | (1:10,000), (1 µL) |
| Antibody | Anti-p24 (rabbit monoclonal) | Sino Biological | Cat # 11695-V08E RRID:N/A | (1:500), (10 µL) |
| Antibody | Anti-rhodopsin/C9 (mouse monoclonal) | Novus | Cat # NBP1-47602 RRID:AB_10010560 | (1:1000), (5 µL) |
| Antibody | IRDye 680RD donkey anti-rabbit | LI-COR | Cat # 926–68073; RRID:AB_10954442 | (1:5000), (1 µL) |
| Antibody | IRDye 800CW donkey anti-mouse | LI-COR | Cat # 926–32212; RRID: AB_621847 | (1:5000), (1 µL) |
| Antibody | Nucleocapsid (Human monoclonal) | Mount Sinai | Clone 1C7C7 RRID:N/A | Isolated by the Center for Therapeutic Antibody Discovery at the Icahn School of Medicine at Mount Sinai (1:1000), (1 µL) |
| Recombinant DNA reagent | pcDNA3.1-SARS2-Spike | Addgene | Cat # 145032 | |
| Recombinant DNA reagent | pcDNA3.1-SARS2-Spike-D614G | This study Addgene | Cat # 166850 | |
| Recombinant DNA reagent | pLentiEGFP | Addgene | Cat # 138152 | |
| Recombinant DNA reagent | pMD2.G | Addgene | Cat # 12259 | |
| Peptide, recombinant protein | Spike protein D614 | Sino Biological | Cat # 40591-V08H | |
| Peptide, recombinant protein | Spike protein G614 | Sino Biological | Cat # 40591-V08H3 | |
| Commercial assay or kit | psPAX2 | Addgene | Cat # 12260 | |
| Commercial assay or kit | Q5 site-directed mutagenesis kit | NEB | Cat # E0554S | |
| Commercial assay or kit | Luna Universal One-step qPCR kit | NEB | Cat # E3005L | |
| Software, algorithm | NextStrain | Hadfield et al., 2018 | https://nextstrain.org/ncov | |
| Software, algorithm | FlowJo v10 | BD Bioscience | ||
| Software, algorithm | GraphPad Prism 8 | GraphPad | ||
| Software, algorithm | Image Lab 6.1 | BioRad | ||
| Software, algorithm | Image J 1.51 | NIH/LOCI | ||
| Software, algorithm | NetMHC 4.0 | Andreatta and Nielsen, 2016 | http://www.cbs.dtu.dk/services/NetMHC/ | |
| Other | SARS-CoV-2 | CDC | Isolate USA-WA1/2020 (NR-52281) | Deposited by the Center for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH |





