Repurposing eflornithine to treat a patient with a rare ODC1 gain-of-function variant disease
Figures
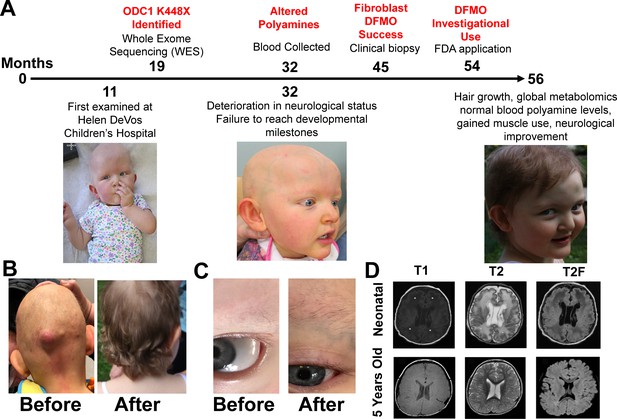
Patient phenotypes and metabolites before and after eflornithine treatment.
Panel A shows the timeline of events for the patient with milestones marked on the top and clinical observations below. Panels B-C show hair growth and muscle tone are the most noticeable phenotype changes with treatment. Follicular cysts recurred on back, neck, and posterior scalp (bottom left images). First hair growth was eyebrows 1 month into treatment (bottom right images). Panel D shows MRI before and after eflornithine treatment. Neonatal: Axial T1 (TR 483 ms, TE 9 ms, and flip angle 63 degrees), T2 (TR 3250 ms, TE 220 ms, and flip angle 90 degrees), and T2-FLAIR (TR 8002 ms, TE 122 ms, and flip angle 90 degrees) show marked abnormal signal of cerebral white matter (*) and several subependymal cysts (arrows). Five years of age: Axial T1 (TR 809 ms, TE 16 ms, and flip angle 111 degrees), T2 (TR 4850 ms, TE 107 ms, and flip angle 142 degrees), and T2-FLAIR (TR 6002 ms, TE 91 ms, and flip angle 90 degrees) show decrease in cerebral white matter volume, but normalization of signal and resolution of subependymal cysts.
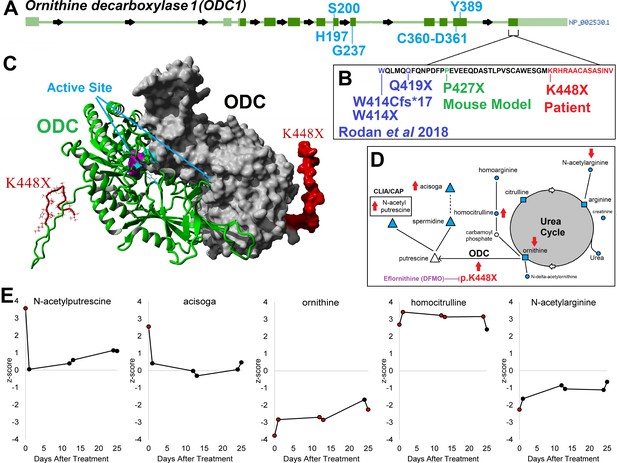
ODC1/ODC clinical variant c.1342 A > T/ p.K448X and support for eflornithine treatment.
Panel A shows the gene structure for ODC1 (ornithine decarboxylase 1) with active site amino acids labeled in blue and the last exon identified. In the last exon cluster, a mouse model variant and four different patient variants including our patient’s K448X variant are described (Panel B, red). The patient variant falls on the disordered C-terminus of ODC, where the two active sites are composed of amino acids from each of two ODC proteins forming a dimer (Panel C). The patient with K448X variant displays alterations of metabolic pathways (Panel D) including polyamines (triangle), urea (square), and others (circle). Metabolites measured are marked in cyan and those altered by K448X with red arrows based on direction of changes seen in the patient. Panel E shows changes in metabolite levels during treatment with eflornithine, with elevated levels of N-acetylputrescine and acisoga decreasing on therapy.
Videos
Treatment progression after 4 months of eflornithine therapy.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Homo sapiens) | ODC1 | NCBI Gene | Gene ID: 4953 | https://www.ncbi.nlm.nih.gov/gene/4953 |
| Chemical compound, drug | Eflornithine (DFMO) | Sanofi Aventis | Supplied for study | https://pubchem.ncbi.nlm.nih.gov/compound/Eflornithine |
| Biological sample (Homo sapiens) | Blood EDTA tubes | Freshly isolated blood from patient | ||
| Software, algorithm | YASARA | YASARA | http://www.yasara.org/ | Protein modelling |
| Commercial assay or kit | Liquid chromatography paired massspectrometry | Metabolon, Morrisville, NC | https://www.metabolon.com/ |
Additional files
-
Source data 1
Global Metabolomics.
- https://cdn.elifesciences.org/articles/67097/elife-67097-data1-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/67097/elife-67097-transrepform-v1.docx





