Rad53 checkpoint kinase regulation of DNA replication fork rate via Mrc1 phosphorylation
Figures
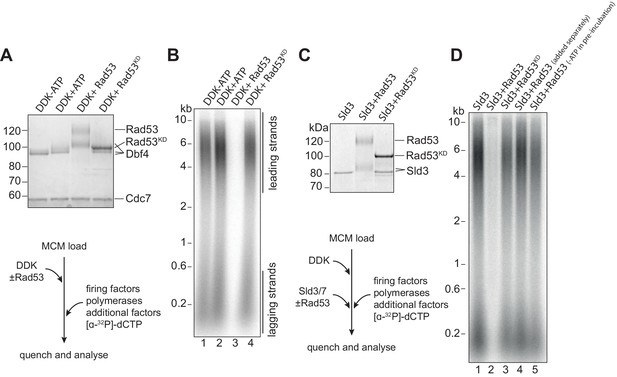
Rad53 inhibition of origin firing.
(A) DDK was incubated with Rad53 or Rad53KD (K227A, K339A) for 15 min, separated by SDS-PAGE, and stained with coomassie. (B) DDK was incubated with Rad53 for 15 min and then added to a standard three-step in vitro replication reaction (see Materials and methods for more details). After 20 min, reactions were stopped with EDTA and products were separated on an alkaline agarose gel. (C) Sld3/7 was incubated with Rad53 as in (A). (D) Sld3/7 was incubated with Rad53 and added to a replication reaction. In lane 4, Sld3/7 and Rad53 were incubated separately from each other prior to addition to the replication reaction, and in lane 5, ATP was omitted during the pre-incubation.
-
Figure 1—source data 1
Original gel images for Figure 1.
- https://cdn.elifesciences.org/articles/69726/elife-69726-fig1-data1-v2.pdf
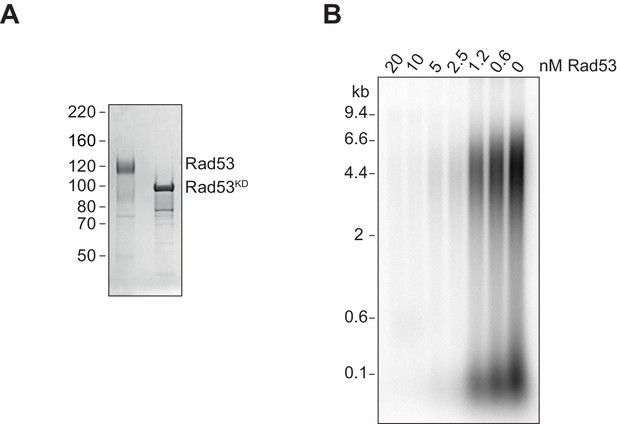
Rad53 phosphorylation of Sld3/7.
(A) Coomassie-stained SDS-PAGE of Rad53 (shifted due to autophosphorylation during expression in bacteria [Gilbert et al., 2001]) and Rad53KD. (B) Replication reactions with Sld3/7 pre-incubated with Rad53 at varying concentrations.
-
Figure 1—figure supplement 1—source data 1
Original gel images for Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/69726/elife-69726-fig1-figsupp1-data1-v2.pdf
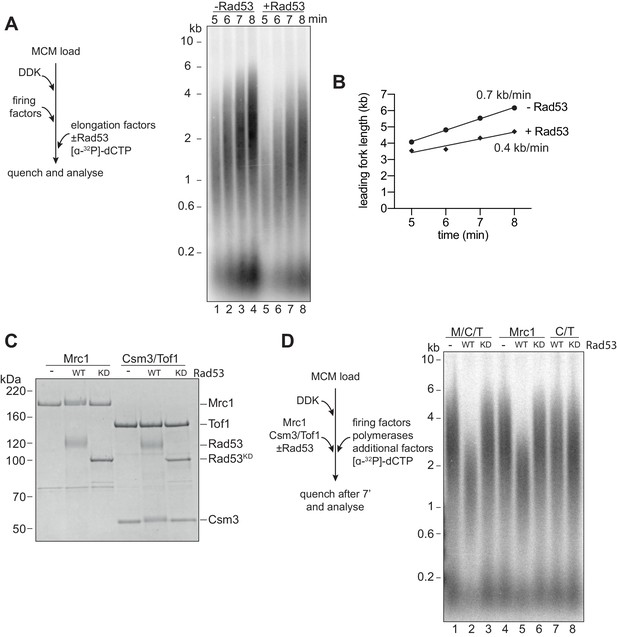
Rad53 inhibition of replication elongation via Mrc1.
(A) Elongation factors (here, defined as RPA, Ctf4, TopoI, Mrc1, Csm3/Tof1, Polɑ, and Mcm10) were pre-incubated with Rad53 prior to addition to a four-step replication reaction that was stopped at the indicated timepoints. (B) Leading fork lengths (see Materials and methods for quantification method) at each timepoint from (A) with a linear fit. (C) Mrc1 or Csm3/Tof1 were incubated with Rad53, separated by SDS-PAGE, and stained with coomassie. (D) M/C/T or individual Mrc1 and Csm3/Tof1 were incubated with Rad53 prior to addition to a replication reaction. Reactions were stopped at 7 min (not completion).
-
Figure 2—source data 1
Original gel images for Figure 2.
- https://cdn.elifesciences.org/articles/69726/elife-69726-fig2-data1-v2.pdf
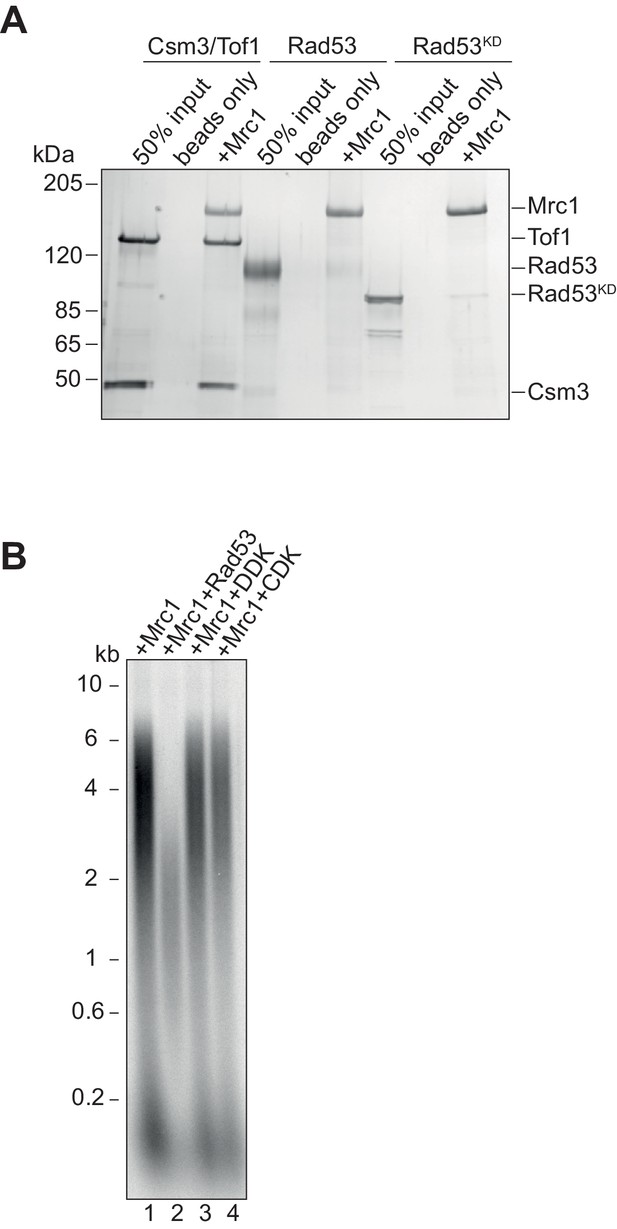
Mrc1 and Rad53 binding.
(A) In vitro co-immunoprecipitation experiments were performed by first incubating anti-flag M2 magnetic beads with Mrc1 and ATP, then adding equimolar amounts of the indicated protein. Samples were washed then eluted from the beads with 3x-flag peptide, separated on SDS-PAGE and coomassie stained. (B) Mrc1 inhibition by replication associated kinases. Mrc1 was incubated with Rad53, DDK, or CDK prior to addition to the replication assay, and reactions were stopped at 7 min.
-
Figure 2—figure supplement 1—source data 1
Original gel images for Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/69726/elife-69726-fig2-figsupp1-data1-v2.pdf
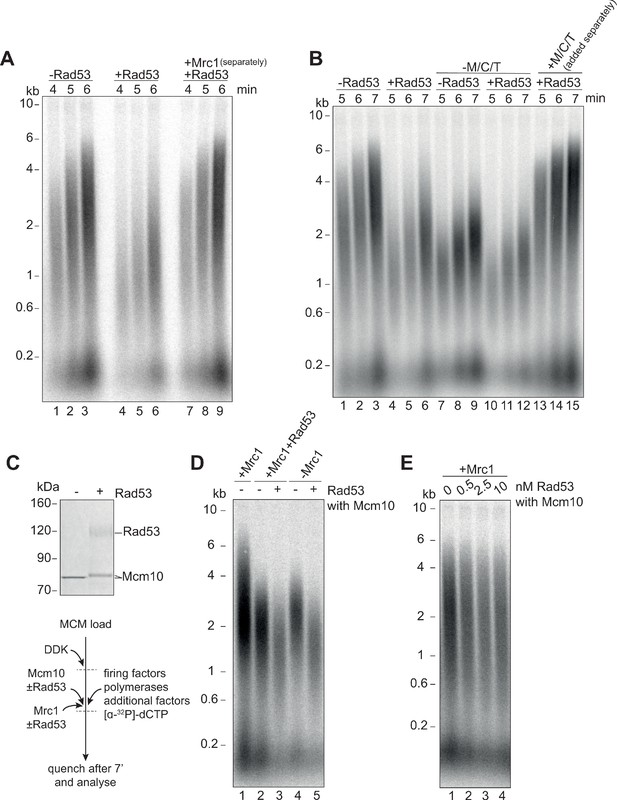
Mrc1 phosphorylation is necessary for Rad53 dependent inhibition of elongation.
(A) Elongation factors were pre-incubated with Rad53 prior to addition to a four-step replication reaction that was stopped at the indicated timepoints. Mrc1 was omitted from the pre-incubation step in lanes 7–9 and added separately. (B) Reactions were performed as in (A) except M/C/T was omitted from the pre-incubation with Rad53 and added separately as indicated. (C) Mcm10 was incubated with Rad53, separated by SDS-PAGE, and stained with coomassie. (D) Mrc1 and Mcm10 were incubated with Rad53 prior to addition to a replication reaction (scheme to the left). Mrc1 was omitted from lanes 4 and 5. Reactions were stopped at 7 min. (E) Mcm10 was incubated with the indicated concentration of Rad53 prior to addition to a replication reaction in the presence of Mrc1.
-
Figure 3—source data 1
Original gel images for Figure 3A and B.
- https://cdn.elifesciences.org/articles/69726/elife-69726-fig3-data1-v2.pdf
-
Figure 3—source data 2
Original gel images for Figure 3C, D and E.
- https://cdn.elifesciences.org/articles/69726/elife-69726-fig3-data2-v2.pdf
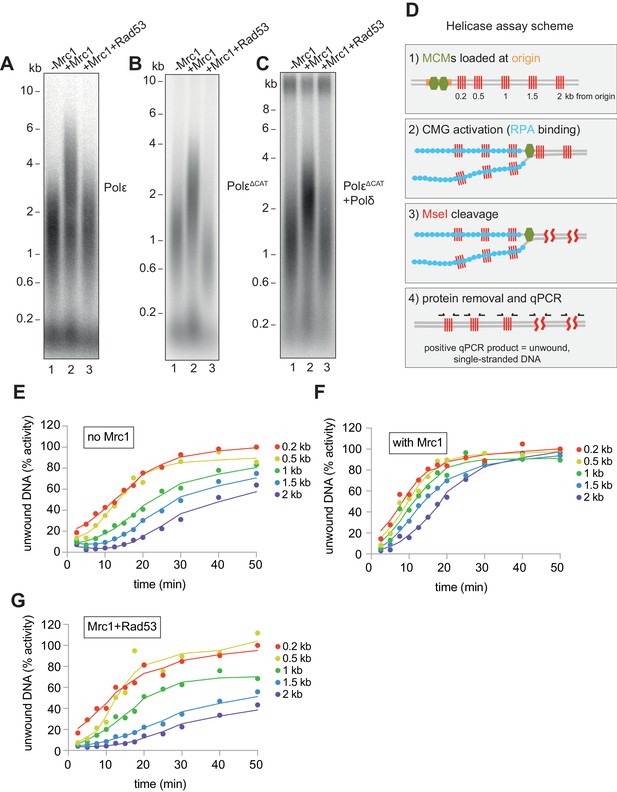
Mrc1 regulation of replication rate.
(A) Mrc1 was omitted, incubated alone, or incubated with Rad53 prior to adding to a replication reaction. Reactions were stopped after 7 min. (B) Reactions as in (A) but using the catalytically-dead mutant (PolεΔCAT) of Polε, and reactions were stopped at 10 min. (C) Reactions as in (A) but with PolεΔCAT and the addition of PCNA, RFC, and Polδ. (D) Helicase assay scheme. After MCMs are loaded specifically at the origin, CMGs are activated and unwind DNA. At each timepoint, MseI is added to digest DNA that is double-stranded; MseI does not digest single-stranded, RPA-coated DNA. The reactions are then quenched, proteins removed, and qPCR is performed using primers flanking the MseI cleavage sites, which generate a signal from the unwound DNA. (E) Timecourse of reactions depicted in (D). Data is normalised to the amount of unwound DNA at the closest MseI site (0.2 kb from the origin) at the last timepoint (see Materials and methods for more detail). (F) Reactions as in (E) with the addition of Mrc1. (D) Reactions as in (E) where Mrc1 is incubated with Rad53.
-
Figure 4—source data 1
Original gel images for Figure 4A and B.
- https://cdn.elifesciences.org/articles/69726/elife-69726-fig4-data1-v2.pdf
-
Figure 4—source data 2
Original gel images for Figure 4C.
- https://cdn.elifesciences.org/articles/69726/elife-69726-fig4-data2-v2.pdf
-
Figure 4—source data 3
Source data for Figure 4E, F and G.
- https://cdn.elifesciences.org/articles/69726/elife-69726-fig4-data3-v2.xlsx
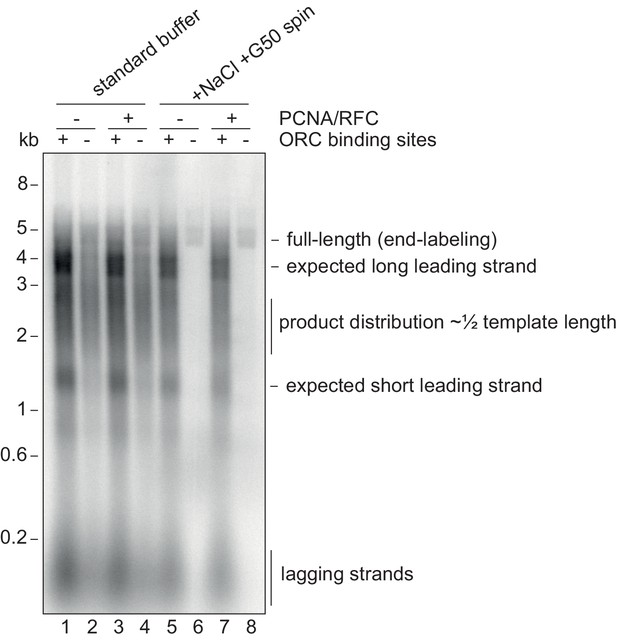
Origin-specific MCM loading conditions.
Replication reactions using 10 nM of the 5 kb CMG helicase assay template (or a version without the ORC binding sites) using normal replication buffer or using the origin-specific loading conditions, which include 80 mM NaCl and processing over a G50 column. The template was linearised prior to the reaction with NheI leading to a 3.5 kb and 1.5 kb distance between the origin and the ends of the template. Note the smear of products centered at 2.5 kb from non-specific loading, which would produce leading strands that average half-template length.
-
Figure 4—figure supplement 1—source data 1
Original gel images for Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/69726/elife-69726-fig4-figsupp1-data1-v2.pdf
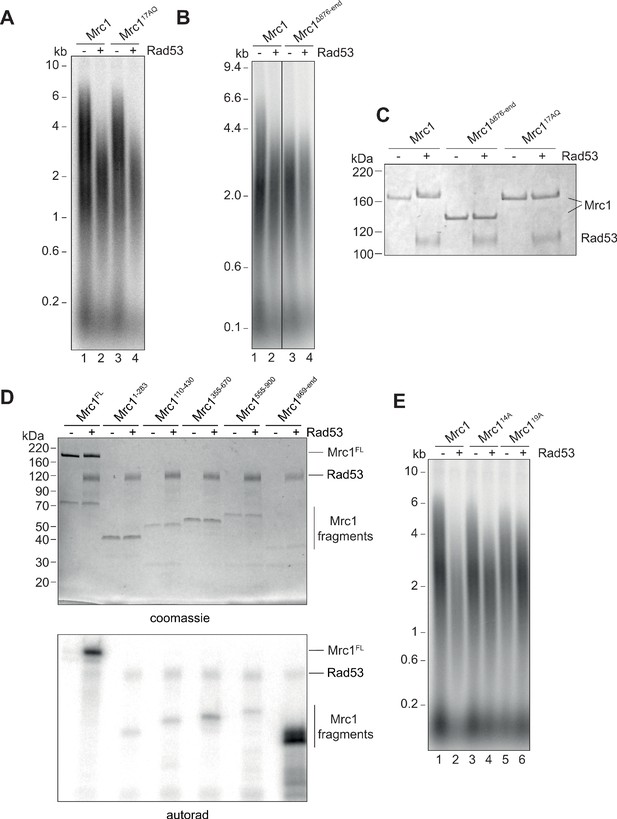
Identification of Rad53-dependent phospho-sites on Mrc1.
(A) Mrc117AQ mutant was incubated with Rad53 prior to addition to in vitro replication for 7 min. (B) C-terminal truncation of Mrc1 was incubated with Rad53 prior to addition to in vitro replication for 7 min. Note that all lanes were run on the same alkaline agarose gel, but other samples between lanes 2 and 3 were removed for clarity. (C) Mrc1, Mrc1 truncation, and Mrc117AQ were incubated with Rad53 and separated by SDS-PAGE then stained with coomassie. (D) Fragments of Mrc1 were incubated with Rad53, separated by SDS-PAGE, then subjected to autoradiography. (E) Mrc1, Mrc114A, and Mrc119A were incubated with Rad53 prior to addition to in vitro replication assay for 7 min.
-
Figure 5—source data 1
Original gel images for Figure 5A, B and C.
- https://cdn.elifesciences.org/articles/69726/elife-69726-fig5-data1-v2.pdf
-
Figure 5—source data 2
Original gel images for Figure 5D and E.
- https://cdn.elifesciences.org/articles/69726/elife-69726-fig5-data2-v2.pdf
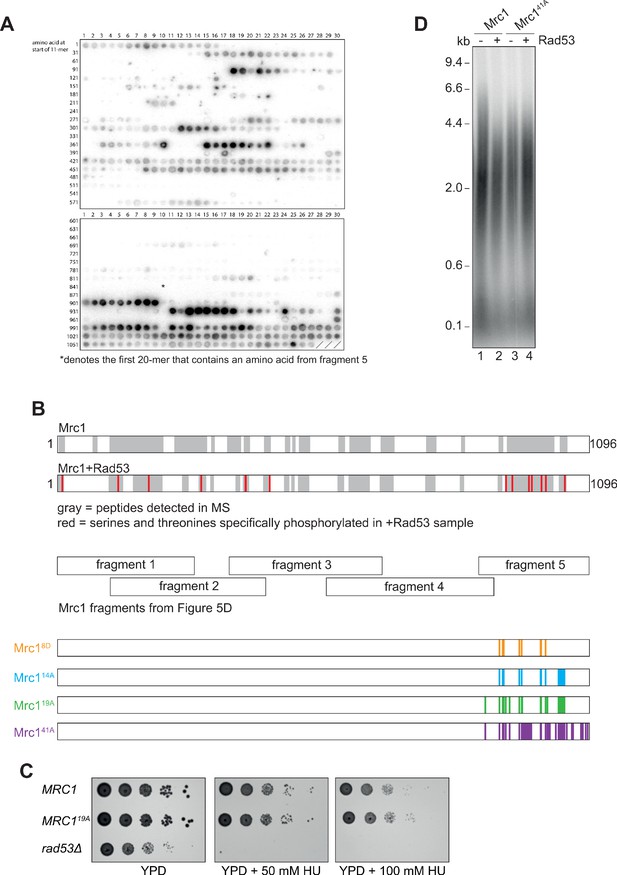
Identification of Rad53-dependent phospho-sites on Mrc1.
(A) Autoradiograph of peptide array of 20-mers scanning the Mrc1 peptide sequence after incubation with Rad53 and γ32P-ATP. The asterisk (*) denotes the first 20-mer that contains an amino acid from fragment 5 of the fragment analysis experiment (Figure 5D). (B) Sites identified by mass spectrometry. The gray highlighting indicates Mrc1 peptides that were detected, and red indicates serines and threonines that were specifically phosphorylated in the Rad53 sample (see Figure 5—figure supplement 1—source data 3). Note the last seven red amino acids fall within fragment 5 of the fragment analysis (Figure 5D). (C) MRC1 (yAWM336), MRC119A (yAWM233), and rad53Δsml1Δ (yAWM338) cells were grown to log-phase and then serially diluted and spotted on the indicated plates. (D) Mrc1 or Mrc141A were incubated with Rad53 prior to addition to a replication reaction for 7 min.
-
Figure 5—figure supplement 1—source data 1
Original gel and plate images for Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/69726/elife-69726-fig5-figsupp1-data1-v2.pdf
-
Figure 5—figure supplement 1—source data 2
List of peptides synthesised for peptide array in Figure 5—figure supplement 1A.
- https://cdn.elifesciences.org/articles/69726/elife-69726-fig5-figsupp1-data2-v2.docx
-
Figure 5—figure supplement 1—source data 3
Source data for Figure 5—figure supplement 1B.
- https://cdn.elifesciences.org/articles/69726/elife-69726-fig5-figsupp1-data3-v2.xlsx
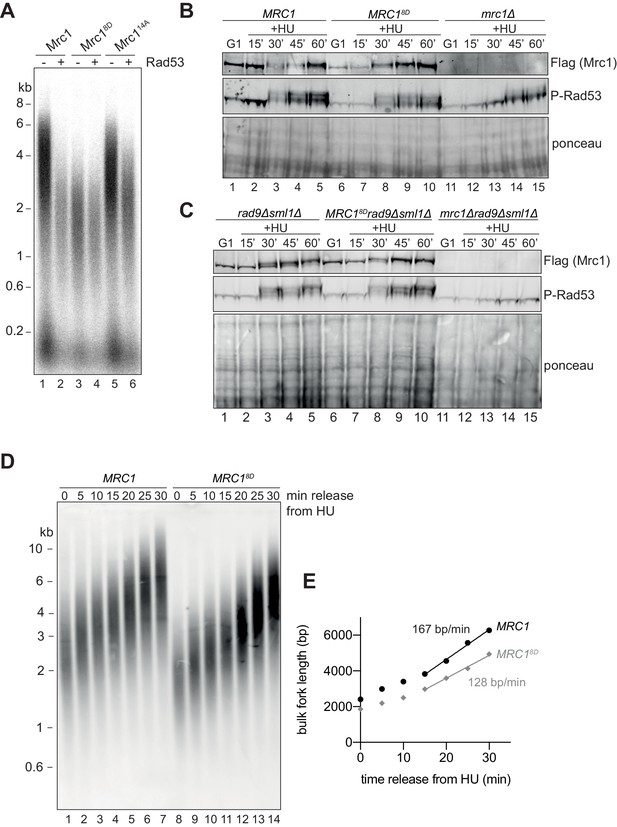
Mrc18D slows fork rate in vitro and in vivo.
(A) Mrc1 and Mrc18D were incubated with Rad53 prior to addition to replication reaction for 7 min. (B) Cells harbouring MRC1-3xFLAG (yAWM336), MRC18D-3xFLAG (yAWM291), or mrc1Δ (yAWM217) were synchronised in G1 with ɑ-factor then released into media with or without 200 mM HU for the indicated timepoints. TCA lysates were then analysed by western blot with the indicated antibodies. (C) Cells that contained sml1Δ rad9Δ in addition to MRC1-3xFLAG (yAWM346), MRC18D-3xFLAG (yAWM348), or mrc1Δ (yJT135) were treated as in (B). (D) Cells harbouring MRC1-3xFLAG (yAWM373) or MRC18D-3xFLAG (yAWM375) with Gal1prom-dmdNK and Gal1prom-hENT were synchronised in G1 with ɑ-factor then released into media with BrdU and HU for 1 hr, the released into media with thymidine to chase the BrdU labelling. Following DNA extraction, samples were separated on an alkaline gel, transferred to a nylon membrane, and immunoblotted with anti-BrdU antibody. (E) quantification of samples from (D) (see Materials and methods for detailed method).
-
Figure 6—source data 1
Original gel images for Figure 6.
- https://cdn.elifesciences.org/articles/69726/elife-69726-fig6-data1-v2.pdf
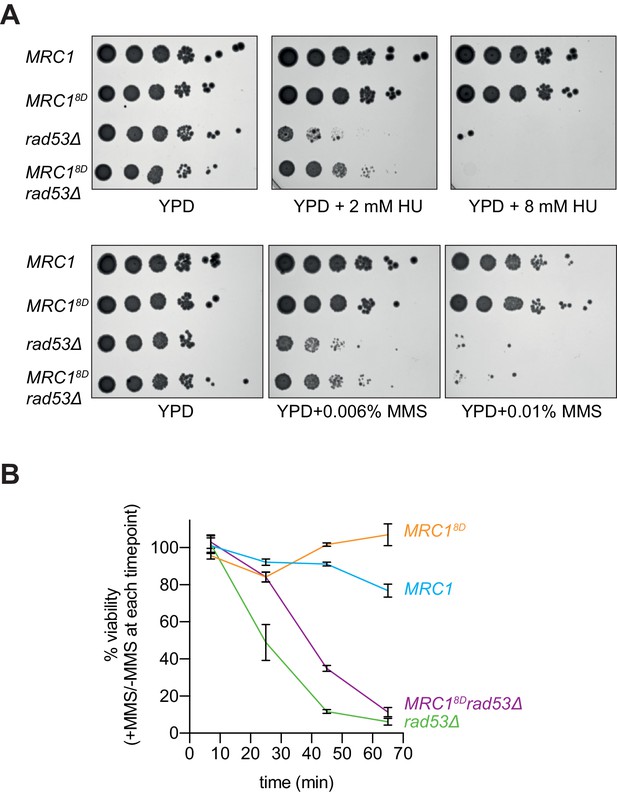
Mrc18D partially rescues rad53Δ sensitivity to replication stress.
(A) Cells that contained sml1Δ as well as MRC1-3xFLAG (yAWM337), MRC18D-3xFLAG (yAWM292), MRC1-3xFLAG and rad53Δ (yAWM338), or MRC18D-3xFLAG and rad53Δ (yAWM293) were spotted as 1:10 serial dilutions onto YPD plates supplemented with the indicated drugs. (B) Cells from (A) were arrested in G1 with alpha-factor then released into media with or without 0.02% MMS for the indicated timepoints then plated on YPD plates in triplicate (mean +/- s.e.m).
-
Figure 7—source data 1
Original plate images for Figure 7A.
- https://cdn.elifesciences.org/articles/69726/elife-69726-fig7-data1-v2.pdf
-
Figure 7—source data 2
Source data for Figure 7B.
- https://cdn.elifesciences.org/articles/69726/elife-69726-fig7-data2-v2.xlsx
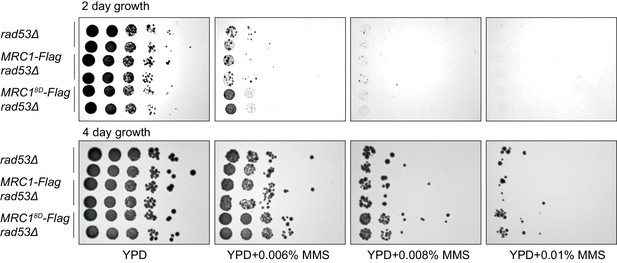
Just after separating tetrad spores, two strains of each of the indicated genotype (with sml1Δ) were identified, grown for 6 hr, and then spotted in 1:10 serial dilutions on the indicated plates.
Note the similar phenotype between the first four rows indicating MRC1-3xFLAG does not alter phenotype.
-
Figure 7—figure supplement 1—source data 1
Original plate images for Figure 7—figure supplement 1.
- https://cdn.elifesciences.org/articles/69726/elife-69726-fig7-figsupp1-data1-v2.pdf
Tables
Protein purification strategy.
| Protein | Purification strategy (see Deegan et al., 2016; Yeeles et al., 2015; Yeeles et al., 2017 for more details) |
|---|---|
| MCM-Cdt1 | Yeast expression, calmodulin pull-down, EGTA elution, gel filtration |
| Cdc6 | Bacterial expression, glutathione pull-down, precission protease elution, HTP column, dialysis |
| ORC | Yeast expression, calmodulin pull-down, EGTA elution, gel filtration |
| DDK | Yeast expression, calmodulin pull-down, EGTA elution, gel filtration |
| Rad53 | Bacterial expression, Ni-NTA pull-down, imidazole elution, gel filtration |
| Mrc1 | Yeast expression, Flag pull-down, flag peptide elution, MonoQ, dialysis |
| Cdc45 | Yeast expression, Flag pull-down, flag peptide elution, HTP column, dialysis |
| Dpb11 | Yeast expression, Flag pull-down, flag peptide elution, gel filtration |
| Polε and Polεexo- | Yeast expression, calmodulin pull-down, EGTA elution, heparin column, gel filtration |
| PolεΔCAT | Yeast expression, calmodulin pull-down, EGTA elution, MonoQ, gel filtration |
| GINS | Bacterial expression, Ni-NTA pull-down, imidazole elution, MonoQ, gel filtration |
| CDK | Yeast expression, calmodulin pull-down, TEV elution, Ni-NTA column, gel filtration |
| RPA | Yeast expression, calmodulin pull-down, EGTA elution, heparin column, gel filtration |
| Ctf4 | Yeast expression, calmodulin pull-down, EGTA elution, MonoQ, gel filtration |
| TopoI | Yeast expression, calmodulin pull-down, EGTA elution, gel filtration |
| Csm3/Tof1 | Yeast expression, calmodulin pull-down, TEV elution, gel filtration |
| Polɑ | Yeast expression, calmodulin pull-down, EGTA elution, MonoQ, gel filtration |
| Sld3/7 | Yeast expression, IgG Sepharose six pull-down, TEV elution, Ni-NTA column, gel filtration |
| Mcm10 | Bacterial expression, Ni-NTA pull-down, imidazole elution, gel filtration |
| Sld2 | Yeast expression, ammonium sulphate precipitation, Flag pull-down, flag peptide elution, SP column, dialysis |
| RFC | Yeast expression, calmodulin pull-down, EGTA elution, MonoS, gel filtration |
| PCNA | Bacterial expression, ammonium sulphate precipitation, SP column, heparin column, DEAE column, MonoQ, gel filtration |
| Polδ | Yeast expression, calmodulin pull-down, EGTA elution, heparin column, gel filtration |
Primers used in CMG helicase assay.
| Site (distance from origin) | Primer numbers | Sequences |
|---|---|---|
| 200 bp | AWM107 AWM109 | CACTGCACCAAGGTAACACTC GAAGTCAGAGCTGGAGAATCCG |
| 500 bp | AWM111 AWM112 | CCCTACTTCAGCGCCATTCG TAACGGAAGCACCGAATCGT |
| 1000 bp | AWM113 AWM115 | CTCGTTGTGACGCCAATCAG ACATTGAGCCTACGCATCTGT |
| 1500 bp | AWM78 AWM79 | ACTACTGTCACTTCTGAGGGTTC CAGAGGGATGCGTAGTCGTG |
| 2000 bp | AWM116 AWM117 | CGGGGGAAGGAACTCTTGC AGGGGTCGTCAAGCAGAGAT |
| control site (not flanking MseI) | AWM84 AWM85 | CTCTGCTTGACGACCCCTTG TGTCCGTCCGAGAGCGATA |
Yeast strains generated in this study.
| Strain | Genotype (all in W303 background) |
|---|---|
| yVP8 | MATa bar1::hygR pep4::kanR his3:pRS303-Gal1prom-3xFLAG-DPB11:HIS3 |
| yVP7 | MATa bar1::hygR pep4::kanR his3:pRS303-Gal1prom-MRC117AQ-3xFLAG:HIS3 |
| yAWM106 | MATa bar1::hygR pep4::kanR his3:pRS303-Gal1prom-MRC11-875-3xFLAG:HIS3 |
| yAWM107 | MATa bar1::hygR pep4::kanR his3:pRS303-Gal1prom-MRC18D-3xFLAG:HIS3 |
| yAWM105 | MATa bar1::hygR pep4::kanR his3:pRS303-Gal1prom-MRC114A-3xFLAG:HIS3 |
| yAWM115 | MATa bar1::hygR pep4::kanR his3:pRS303-Gal1prom-MRC119A-3xFLAG:HIS3:NAT |
| yAWM108 | MATa bar1::hygR pep4::kanR his3:pRS303-Gal1prom-MRC141A-3xFLAG:HIS3 |
| yAWM343 | MATa/ɑ Mrc18D-3xFLAG:natR/MRC1 sml1::kanR/SML1 rad9::LEU2/RAD9 |
| yAWM373 | MATa MRC1-3xFLAG:natR leu2: Gal1prom-hENT:LEU2 trp1: Gal1prom-dmdNK:TRP1 |
| yAWM375 | MATa MRC18D-3xFLAG:natR leu2: Gal1prom-hENT:LEU2 trp1: Gal1prom-dmdNK:TRP1 |
| yAWM337 | MATa MRC1-3xFLAG:natR sml1::kanR |
| yAWM292 | MATa MRC18D-3xFLAG:natR sml1::kanR |
| yAWM338 | MATa MRC1-3xFLAG:natR rad53::LEU2 sml1::kanR |
| yAWM293 | MATa MRC18D-3xFLAG:natR rad53::LEU2 sml1::kanR |
| yAWM336 | MATa MRC1-3xFLAG:natR |
| yAWM233 | MATa MRC119A:natR |
| yAWM291 | MATa MRC18D-3xFLAG:natR |
| yAWM217 | MATa mrc1::kanR |
| yAWM346 | MATa MRC1-3xFLAG:natR rad9::LEU2 sml1::kanR |
| yAWM348 | MATa MRC18D-3xFLAG:natR rad9::LEU2 sml1::kanR |
| yJT135 | MATa sml1::URA3 rad9::TRP1 mrc1::HIS3 |
DNA plasmids generated in this study.
| Plasmid | Description *see details in the construction notes |
|---|---|
| pAWM7 | pET21b-RAD53K227A,D339A-6xHis |
| pAWM10 | pET21b-MRC11-283-6xHis |
| pAWM11 | pET21b-MRC1110-430-6xHis |
| pAWM12 | pET21b-MRC1355-670-6xHis |
| pAWM13 | pET21b-MRC1555-900-6xHis |
| pAWM14 | pET21b-MRC1869-1096-6xHis |
| pAWM16 | pRS303-GAL1prom-MRC1-3xFLAG, GAL4 |
| pVP14 | pRS303-GAL1prom-MRC117AQ-3xFLAG, GAL4 |
| pAWM35 | pRS303-GAL1prom-MRC11-875-3xFLAG, GAL4 |
| pAWM15 | pRS303-GAL1prom-MRC114A-3xFLAG, GAL4 |
| pAWM25 | pRS40N- MRC119A-3xFLAG (N-terminal truncation for integration) |
| pAWM18 | pRS303-GAL1prom-MRC141A-3xFLAG, GAL4 |
| pAWM17 | pRS303-GAL1prom-MRC18D-3xFLAG, GAL4 |
| pAWM48 | pRS40N-MRC1-3xFLAG (N-terminal truncation for integration) |
| pAWM47 | pRS40N-MRC18D-3xFLAG (N-terminal truncation for integration) |
| pAWM36 | pBS-based template for CMG helicase assay |
| pAWM37 | pBS-based template for CMG helicase assay (no origin) |





