DNA-PK promotes DNA end resection at DNA double strand breaks in G0 cells
Figures
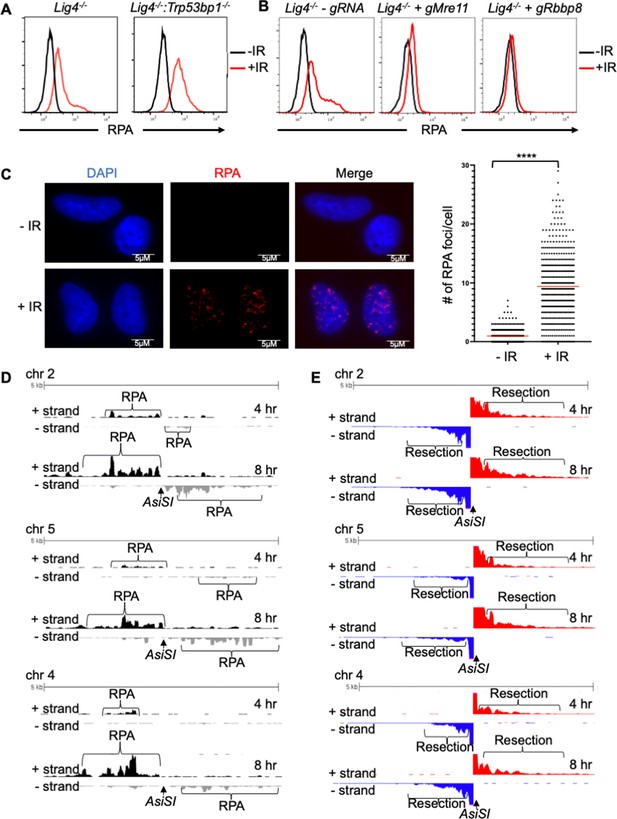
RPA is loaded onto ssDNA after DSBs in G0 mammalian cells.
(A) Flow cytometric analysis of chromatin-bound RPA in G0-arrested Lig4-/- and Lig4-/-:Trp53bp1-/- abl pre-B cells before and 3 hr after 20 Gray IR. Representative of three independent experiments. (B) Flow cytometric analysis of chromatin-bound RPA before and 2 hr after 15 Gy IR in G0-arrested Lig4-/- abl pre-B cells (left), Lig4-/- cells depleted of MRE11 (middle), and Lig4-/- cells depleted of CtIP (right). Representative of three independent experiments. (C) Representative images and quantification of IR-induced RPA foci from three independent experiments in G0-arrested MCF10A cells before and 3 hr after 10 Gray IR. n=365 cells in -IR and n=433 cells in +IR. Red bars indicate average number of RPA foci in - IR = 0.96 and average number of RPA foci in +IR = 9.4 (****p<0.0001, unpaired t test). (D) RPA ChIP-seq tracks at AsiSI DSBs on chromosome 2, 5, and 4 at 4 hr (top) and 8 hr (bottom) after AsiSI endonuclease induction in G0-arrested Lig4-/- abl pre-B cells. (E) Representative END-Seq tracks showing resection at AsiSI DSBs at chromosome 2, 5, and 4 at 4 hr (top) and 8 hr (bottom) after AsiSI induction in G0-arrested Lig4-/- abl pre-B cells. END-seq data is representative from two independent experiments.
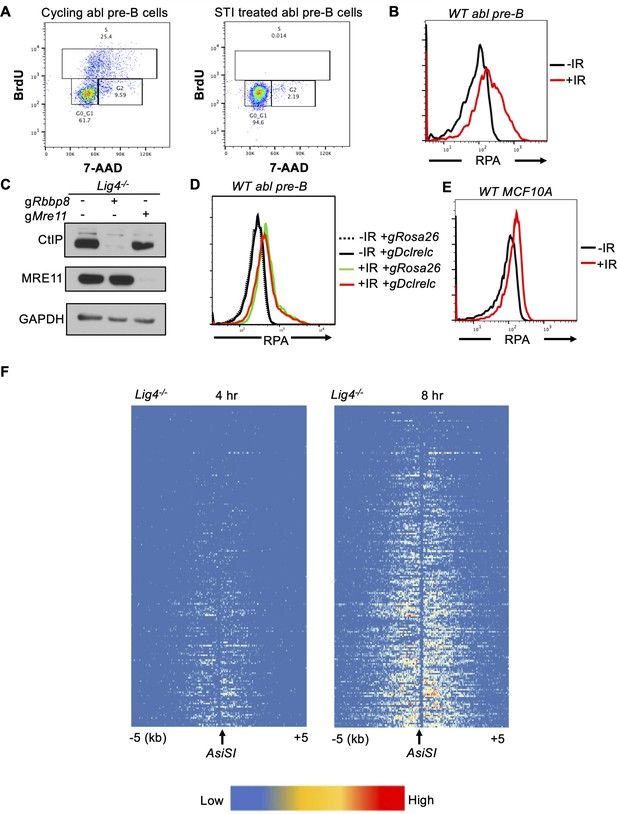
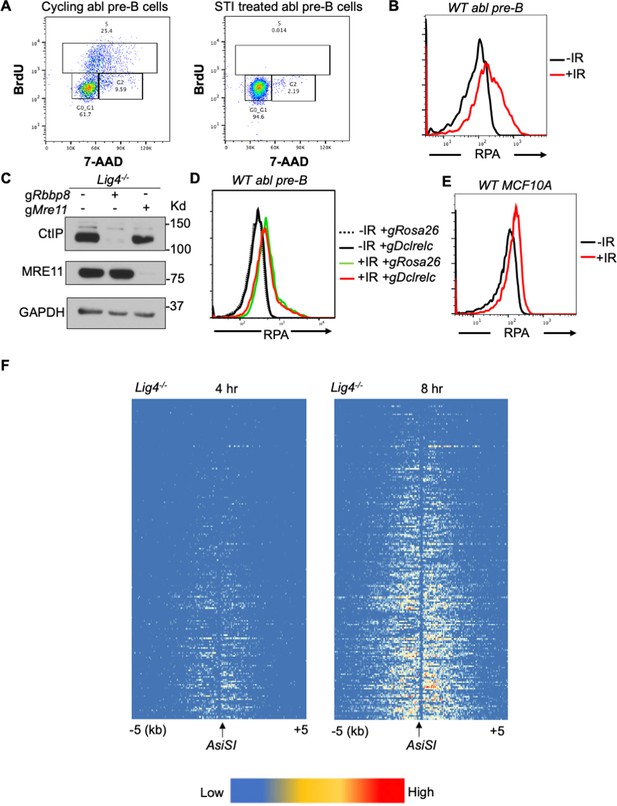
RPA is loaded onto ssDNA after DSBs in G0 mammalian cells.
(A) Flow cytometric analysis of cycling and STI treated Lig4-/- abl pre-B cells for BrdU content (y-axis) and DNA content (7-AAD, x-axis). (B) Flow cytometric analysis of chromatin-bound RPA in wild-type G0-arrested abl pre-B cells before and 3 hr after 20 Gray IR. (C) Western blot of bulk CtIP and MRE11 knockout in Lig4-/- abl pre-B cells. (D) Flow cytometric analysis of chromatin-bound RPA in wild-type G0-arrested abl pre-B cells before and 3 hr after 20 Gray IR in cells depleted of Rosa26 (control) and Artemis, which is encoded by the Dclrelc gene. (E) Flow cytometric analysis of chromatin-bound RPA loading in wild-type G0-arrested MCF10A cells before and 3 hr after 20 Gray IR. (F) Heat maps of RPA ChIP-seq results at top 200 AsiSI sites in G0-arrested Lig4-/- abl pre-B cells 4 hr (left) and 8 hr (right) after AsiSI-endonuclease induction.
-
Figure 1—figure supplement 1—source data 1
Original western blots for Figure 1.
- https://cdn.elifesciences.org/articles/74700/elife-74700-fig1-figsupp1-data1-v2.pdf
The vast majority of AsiSI sites that are cleaved in G0 cells are in close proximity to the transcription start site of actively transcribed genes.
(A) Frequency of broken AsiSI sites versus distance from transcription start sites (TSS). (B) Percentage of broken AsiSI sites within 2000bp of transcription start sites (TSS) and within transcriptionally active genes. (C) Proximity to TSS and level of gene expression for each of the AsiSI sites efficiently cleaved in G0 cells, from which the pie charts in (B) were generated.
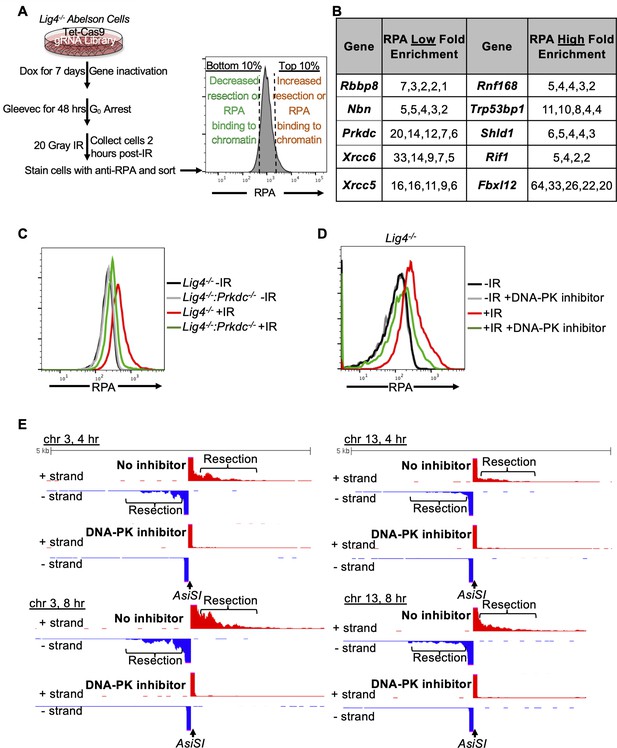
A genome-wide gRNA screen identifies DNA-PK as a factor that promotes DNA end resection in G0.
(A) Schematic of a genome-wide gRNA screen for factors promoting (bottom 10%/RPA low) or inhibiting (top 10%/RPA high) chromatin-bound RPA loading 2 hr after 20 Gray IR in G0-arrested Lig4-/- abl pre-B cells. (B) Fold enrichment of selected gRNAs in low RPA and high RPA populations. Fold enrichment was calculated as the ratio of normalized read number of gRNAs in the low RPA population and that in the high RPA population and vice versa (n=1). (C) Flow cytometric analysis of chromatin-bound RPA in G0-arrested Lig4-/- and Lig4-/-:Prkdc-/- abl pre-B cells before and 3 hr after 15 Gray IR. Data is representative of three independent experiments in two different cell lines. (D) Flow cytometric analysis of chromatin-bound RPA in G0-arrested Lig4-/- abl pre-B cells with and without 10 μM NU7441 (DNA-PK inhibitor) pre-treatment 1 hr before 20 Gray IR. Data is representative of three independent experiments in two different cell lines. (E) Representative END-seq tracks at chromosome 3 (left) and chromosome 13 (right) in G0-arrested Lig4-/- abl pre-B cells 4 hr (top) and 8 hr (bottom) after AsiSI DSB induction, with and without 10 μM NU7441 treatment.
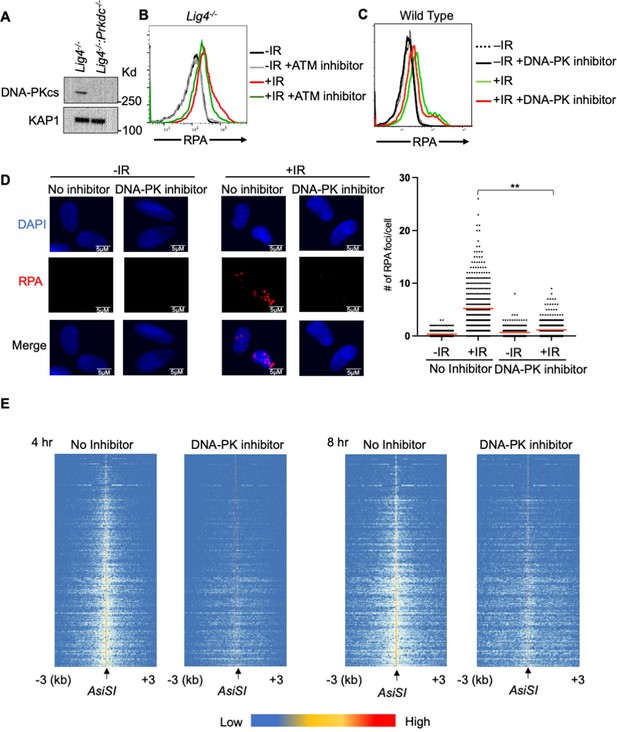
A genome-wide gRNA screen identifies DNA-PK as a factor that promotes DNA end resection in G0.
(A) Western blot analysis of DNA-PKcs protein in Lig4-/- and Lig4-/-:Prkdc-/- abl pre-B cells. (B) Flow cytometric analysis of chromatin-bound RPA in G0-arrested abl pre-B cells with and without 1 hour pre-treatment with 15 μM KU-55933 (ATM inhibitor), before and 3 hr after 20 Gray IR. (C) Flow cytometric analysis of chromatin-bound RPA in G0-arrested abl pre-B cells with and without 1 hr pre-treatment with 10 μM NU7441 (DNA-PK inhibitor), before and 3 hr after 20 Gray IR. (D) Representative images and quantitation from three independent experiments of IR-induced RPA foci in G0-arrested MCF10A cells with and without 10 μM NU7441, before and 3 hr after 10 Gray IR. For No Inhibitor -IR condition, n=426, average number of RPA foci = 0.34. For No Inhibitor +IR condition, n=389, average number of RPA foci = 5.2. For DNA-PK inhibitor treated -IR condition, n=266, average number of RPA foci = 0.66. For DNA-PK inhibitor treated +IR condition, n=441, average number of RPA foci = 1.13. Red bar indicates mean number of RPA foci (**p=0.003). (E) Heat maps of END-seq at top 200 AsiSI DSBs with and without 10 μM NU7441 treatment in G0-arrested Lig4-/- abl pre-B cells 4 hr (left) and 8 hr (right) after AsiSI DSB induction.
-
Figure 2—figure supplement 1—source data 1
Original western blots for Figure 2.
- https://cdn.elifesciences.org/articles/74700/elife-74700-fig2-figsupp1-data1-v2.pdf
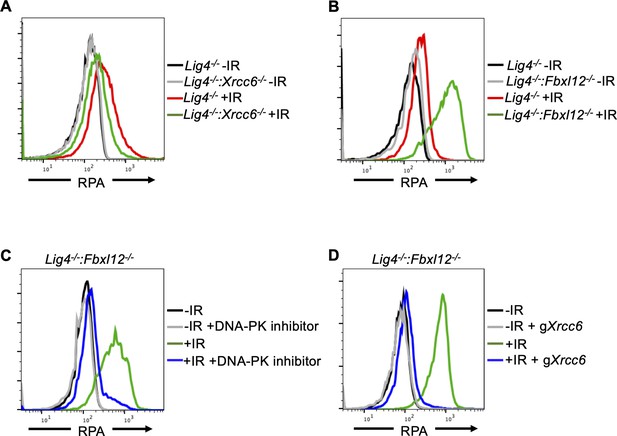
FBXL12 inhibits KU70/KU80-promoted DNA end resection.
(A) Flow cytometric analysis of chromatin-bound RPA in G0-arrested Lig4-/- abl pre-B cells and Lig4-/-:Xrcc6-/- abl pre-B cells before and 3 hr after 20 Gray IR. Data is representative of three independent experiments in two different cell lines. (B) As in A, in G0-arrested Lig4-/- and Lig4-/-:Fbxl12-/- abl pre-B cells. Data is representative of three independent experiments in at least two different cell lines. (C) Flow cytometric analysis of chromatin-bound RPA in G0-arrested Lig4-/-:Fbxl12-/- abl pre-B cells with and without 10 μM NU7441 treatment, before and 3 hr after 20 Gray IR. Data is representative of three independent experiments in at least two different cell lines (D) Flow cytometric analysis of chromatin-bound RPA in G0-arrested Lig4-/-:Fbxl12-/- abl pre-B cells before and after Xrcc6 knockout, before and 3 hr after 15 Gray IR. Data is representative of three independent experiments.
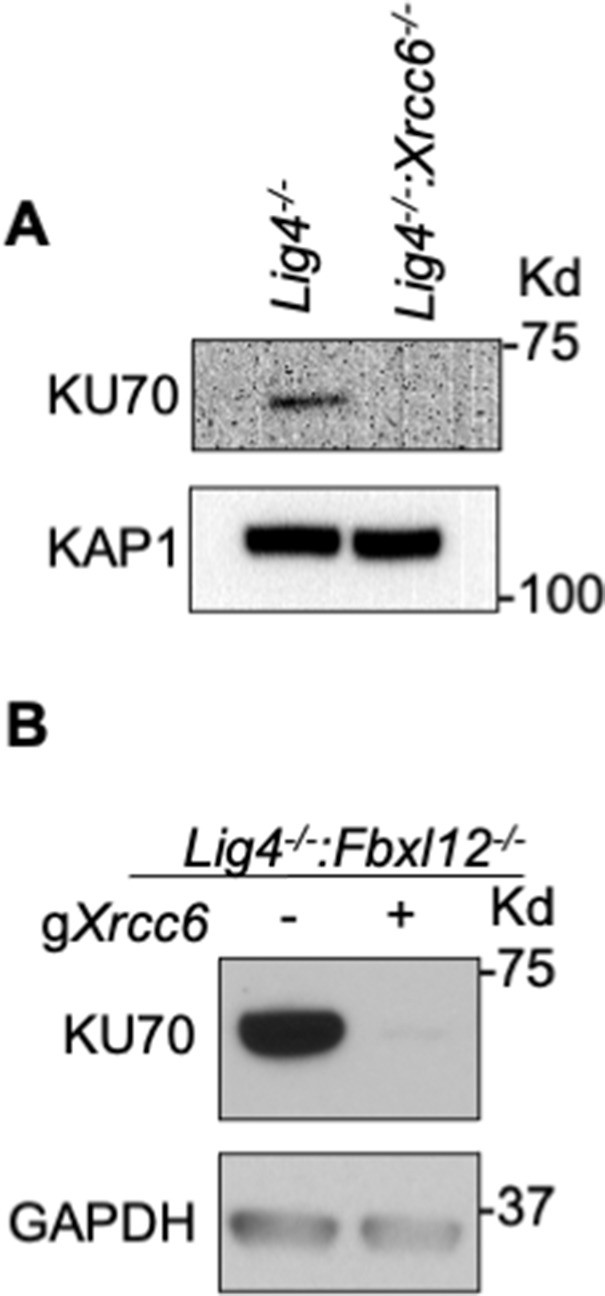
FBXL12 inhibits KU70/KU80-promoted DNA end resection.
(A) Western blot analysis of KU70 protein in Lig4-/- and Lig4-/-:Xrcc6-/- abl pre-B cells. (B) Western blot analysis of KU70 protein in Lig4-/-:Fbxl12-/- abl pre-B cells.
-
Figure 3—figure supplement 1—source data 1
Original western blots showing Ku70 depletion in Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/74700/elife-74700-fig3-figsupp1-data1-v2.pdf
-
Figure 3—figure supplement 1—source data 2
Original western blots for Ku70 depletion in Fbxl12- cells.
- https://cdn.elifesciences.org/articles/74700/elife-74700-fig3-figsupp1-data2-v2.pdf
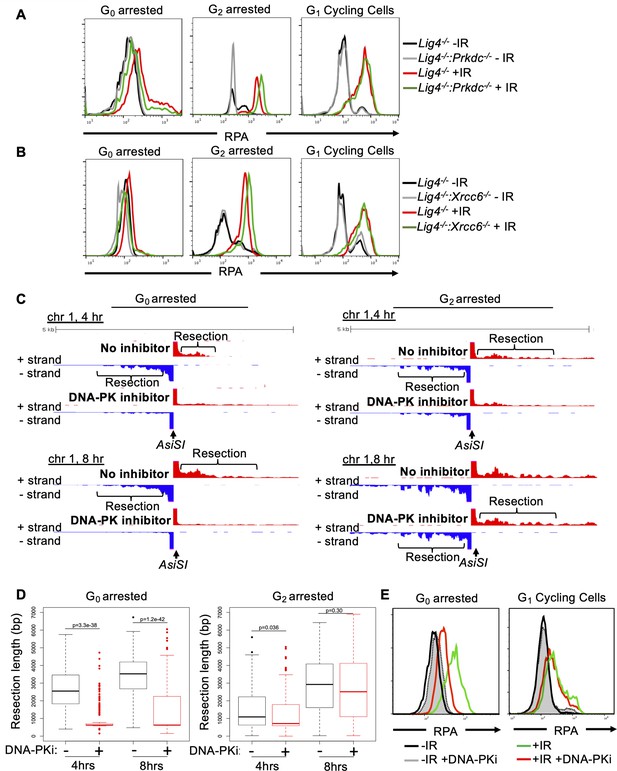
DNA-PK mediates DNA end resection in G0 but not in G1 or G2 phases of the cell cycle.
(A) Flow cytometric analysis of chromatin-bound RPA in Lig4-/- and Lig4-/-:Prkdc-/- abl pre-B cells arrested in G0 (left), arrested in G2 by 10 μM RO-3306 treatment for 16 hr and gated on 4 N (middle), and G1 cells gated on 2 N DNA content in cycling cells (right), before and 3 hr after 20 Gray IR. Data is representative of three independent experiments in at least two different cell lines. (B) As in A in Lig4-/- and Lig4-/-:Xrcc6-/- abl pre-B cells. (C) Representative END-seq tracks in G0 (left) and G2-arrested (right, by 10 μM RO-3306 treatment for 16 hr) Lig4-/- abl pre-B cells, with and without 10 μM NU7441 treatment on chromosome 1, 4 hr (top) and 8 hr (bottom) after AsiSI endonuclease induction. (D) Average resection length in G0-arrested Lig4-/- abl pre-B (left) and G2-arrested Lig4-/- abl pre-B (right) 4 and 8 hr after AsiSI DSB induction, with and without 10 μM NU7441 treatment (DNA-PKi). (E) Flow cytometric analysis of chromatin-bound RPA 4 hr after 20 Gray IR in MCF10A cells arrested in G0 after EGF withdrawal for 48 hr or cycling cells gated on 2 N DNA content, with and without 10 μM NU7441 treatment (DNA-PKi).
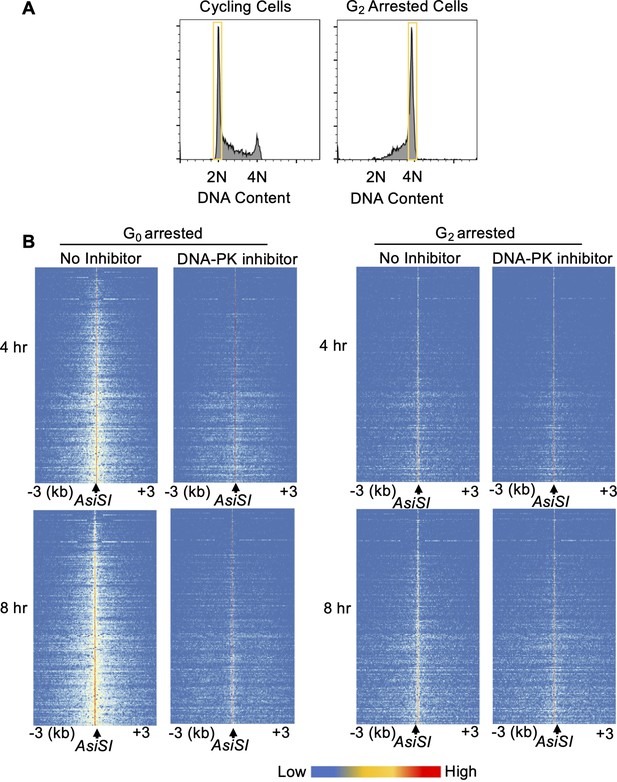
DNA-PK mediates DNA end resection in G0 but not G2.
(A) Flow cytometric analysis of DNA content (7-AAD) in RO-3306 treated (right) and cycling cells (left) showing gating used for RPA flow cytometry analysis. (B) Heat maps of END-seq at top 200 AsiSI DSBs in G0-arrested (left) and G2-arrested (right) Lig4-/- abl pre-B 4 hr (top) and 8 hr (bottom) after AsiSI DSB induction, with and without 10 μM NU7441 treatment.
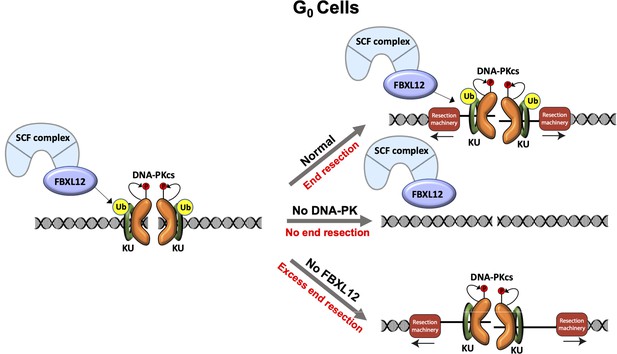
Model of DNA-PK-mediated DNA end resection in G0 cells.
Normally in G0 phase at DSBs, the DNA-PK complex promotes DNA end resection. This resection is counteracted by FBXL12. Without DNA-PK, there is no DNA end resection in G0. Without FBXL12, DNA-PK persists at DSBs and leads to more extensive DNA end resection.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Anti-CtIP (Rabbit polyclonal) | N/A | custom made (Richard Baer, Columbia University) | WB (1:1000) |
| Antibody | Anti-MRE11 (Rabbit polyclonal) | Novus Biologicals | NB100-142 RRID:AB_1109376 | WB (1:2000) |
| Antibody | Anti-GAPDH (GAPDH-71.1) (Mouse monoclonal) | Millipore Sigma | G8795 RRID:AB_1078991 | WB (1:10000) |
| Antibody | Anti-KAP1 (N3C2) (Rabbit polyclonal) | Genetex | GTX102226 RRID:AB_2037324 | WB (1:2000) |
| Antibody | Anti-RPA32 (4E4) (Rat monoclonal) | Cell Signaling Technology | 2,208 S RRID:AB_2238543 | WB (1:1000) FC (1:200) IF (1:500) |
| Antibody | Anti-KU70 (D10A7) (Rabbit monoclonal) | Cell Signaling Technology | 4,588 S RRID:AB_11179211 | WB (1:1000) |
| Antibody | Anti-DNA-PK (SC57-08) (Rabbit monoclonal) | Invitrogen | MA5-32192 RRID:AB_2809479 | WB (1:1000) |
| Antibody | Anti-RPA32 (rabbit polyclonal) | Abcam | ab10359 RRID:AB_297095 | ChIP (10 ug) |
| Antibody | HRP, goat anti-mouse (goat polyclonal) | Promega | W4021 RRID:AB_430834 | WB (1:5000) |
| Antibody | HRP, goat anti-rabbit IgG (goat polyclonal) | Promega | W4011 RRID:AB_430833 | WB (1:5000) |
| Antibody | Alexa Fluor 488, goat anti-rat IgG (goat polyclonal) | BioLegend | 405,418 RRID:AB_2563120 | FC (1:500) |
| Antibody | Alexa Fluor 647, goat anti-rat IgG (goat polyclonal) | BioLegend | 405,416 RRID:AB_2562967 | FC (1:500) |
| Antibody | Alexa Fluor 594, goat anti-rat IgG (goat polyclonal) | BioLegend | 405,422 RRID:AB_2563301 | IF (1:500) |
| Recombinant DNA reagent | pCW-Cas9 (plasmid) | Addgene | 50,661 RRID:Addgene_50661 | |
| Recombinant DNA reagent | pKLV-U6 gRNA(BbsI)-PGKpuro-2ABFP (plasmid) | Addgene | 50,946 RRID:Addgene_50946 | |
| Recombinant DNA reagent | Genome-wide CRISPR guide RNA library V2 (plasmid) | Addgene | 67,988 RRID:Addgene_67988 | |
| Cell line (H. sapiens) | MCF10A | ATCC | CRL-10317 RRID:CVCL_0598 | |
| Cell line (H. sapiens) | MCF10A: iCas9 | This study | Clone 25 | Available upon request |
| Cell line (M. musculus) | WT:iCas9 abl pre-B cells | This study | M63.1.MG36.iCas9.302 | Available upon request |
| Cell line (M. musculus) | Lig4-/-:iCas9 abl pre-B cells | This study | A5.83.MG9.iCas9.16 | Available upon request |
| Cell line (M. musculus) | Lig4-/-:iCas9 abl pre-B cells | This study | A5.115.iCas9.72 | Available upon request |
| Cell line (M musculus) | Lig4-/-:Trp53bp1:iCas9 abl pre-B cells | This study | Clone 82 | Available upon request |
| Cell line (M musculus) | Lig4-/-:Xrcc6-/-:iCas9 abl pre-B cells | This study | Clones 134 and 140 | Available upon request |
| Cell line (M. musculus) | Lig4-/-:Prkdc-/-:iCas9 abl pre-B cells | This study | Clone 6 | Available upon request |
| Cell line (M. musculus) | Lig4-/-:Fbxl12-/-:iCas9 abl pre-B cells | This study | Clone 6 | Available upon request |
| Cell line (M. musculus) | Lig4-/-:iAsiSI abl pre-B cells | This study | Clone 20 | Available upon request |
| Chemical compound, drug | Imatinib | Selleckchem | S2475 | |
| Chemical compound, drug | Doxycycline | Sigma-Aldrich | D9891 | |
| Chemical compound, drug | Polybrene | Sigma Aldrich | S2667 | |
| Chemical compound, drug | Lipofectamine 2000 | Thermo Fisher Scientific | 11668019 | |
| Chemical compound, drug | NU7441 | Selleck Chemicals | S2638 | |
| Chemical compound, drug | KU-55933 | Selleck Chemicals | S1092 | |
| Chemical compound, drug | EGF | PeproTech | AF-100–15 | |
| Chemical compound, drug | Hydrocortisone | Sigma-Aldrich | H-0888 | |
| Chemical compound, drug | Cholera Toxin | Sigma-Aldrich | C-8052 | |
| Chemical compound, drug | Insulin | Sigma-Aldrich | I-1882 | |
| Commercial assay, kit | 7-AAD (DNA stain) | BD Biosciences | 559,925 RRID:AB_2869266 | |
| Commercial assay, kit | Cytofix/Cytoperm solution | BD Biosciences | 554,722 RRID:AB_2869010 | |
| Commercial assay, kit | Perm/Wash Buffer | BD Biosciences | 554,723 RRID:AB_2869011 | |
| Commercial assay, kit | FITC BrdU Flow Kit | BD Biosciences | 559,619 RRID:AB_2617060 | |
| Sequence-based reagent | pKLV lib330F | This study designed based on [Tzelepis et al., 2016] | PCR primers | AATGGACTATCATATGCTTACCGT |
| Sequence-based reagent | pKLV lib490R | This study designed based on Tzelepis et al., 2016 | PCR primers | CCTACCGGTGGATGTGGAATG |
| Sequence-based reagent | PE.P5_pKLV lib195 Fwd | This study designed based on Tzelepis et al., 2016 and standard Illumina adaptor sequences | PCR primers | AATGATACGGCGACCACCGAGATCTGG CTTTATATATCTTGTGGAAAGGAC |
| Sequence-based reagent | P7 index180 Rev | This study designed based on Tzelepis et al., 2016 and standard Illumina adaptor sequences | PCR primers | CAAGCAGAAGACGGCATACGAGAT INDEXGTGACTGGAGTTCAGACGTG TGCTCTTCCGATCCAGACTGCCTTGGGAAAAGC |
| Sequence-based reagent | BU1 | Canela et al., 2016 | PCR primers | 5′-Phos-GATCGGAAGAGCGTCGT GTAGGGAAAGAGTGUU[Biotin-dT]U [Biotin-dT]UUACACTCTTTC CCTACA CGACGCTCTTCCGATC* T-3′ [*phosphorothioate bond] |
| Sequence-based reagent | BU2 | Canela et al., 2016 | PCR primers | 5′-Phos-GATCGGAAGAGCACACG TCUUUUUUUUAGACGTGTGCTC TTCCGATC*T-3′ [*phosphorothioate bond] |
| Sequence-based reagent | Trp53bp1 gRNA sequence | Sequence is from Tzelepis et al., 2016 | N/A | GAACCTGTCAGACCCGATC |
| Sequence-based reagent | Rbbp8 gRNA sequence | Sequence is from Tzelepis et al., 2016 | N/A | ATTAACCGGCTACGAAAGA |
| Sequence-based reagent | Mre11 gRNA sequence | Sequence is from Tzelepis et al., 2016 | N/A | TGCCGTGGATACTAAATAC |
| Sequence-based reagent | Prkdc gRNA sequence | Sequence is from Tzelepis et al., 2016 | N/A | ATGCGTCTTAGGTGATCGA |
| Sequence-based reagent | Xrcc6 gRNA sequence | Sequence is from Tzelepis et al., 2016 | N/A | CCGAGACACGGTTGGCCAT |
| Sequence-based reagent | Fbxl12 gRNA sequence | Sequence is from Tzelepis et al., 2016 | N/A | TTCGCGATGAGCATCTGCA |
| Software, algorithm | Image J | NIH | RRID:SCR_003070 | |
| Software, algorithm | FlowJo | FlowJo | RRID:SCR_008520 | |
| Software, algorithm | Prism | GraphPad | RRID:SCR_002798 | |
| Software, algorithm | Gen5 | Biotek Instruments | RRID:SCR_017317 | |
| Software, algorithm | SeqKit | Shen et al., 2016 | RRID:SCR_018926 | |
| Software, algorithm | Bowtie | Langmead et al., 2009 | RRID:SCR_005476 | |
| Software, algorithm | SAMtools | Li et al., 2009 | RRID:SCR_002105 | |
| Software, algorithm | BEDtools | Quinlan and Hall, 2010 | RRID:SCR_006646 | |
| Other | LSRII Flow cytometer | BD Bioscience | RRID:SCR_002159 | Flow cytometer |
| Other | FACS Celesta Flow Cytometer | BD Bioscience | RRID:SCR_019597 | Flow cytometer |
| Other | FACSAria II Cell Sorter | BD Bioscience | RRID:SCR_018934 | Flow assisted cell sorter |
| Other | Lionheart LX automated microscope | BioTex Instruments | RRID:SCR_019745 | Automated microscope |
| Other | 4-D Amaxa Nucleofecter | Lonza | NA | Nucleofector |





