Evolution of multiple omics approaches to define pathophysiology of pediatric acute respiratory distress syndrome
Figures
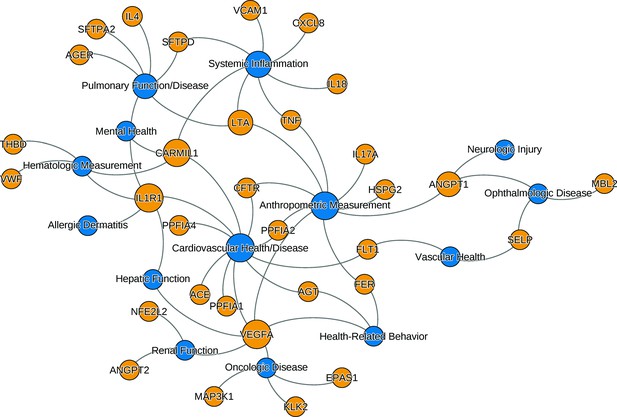
The relationships between ARDS candidate genes and phenotypes reported by genome-wide association studies.
The nodes (circles) represent genes (light gray, gene symbols not italicized) or phenotypes (dark gray), and the associated genotype–phenotype pairs are connected by curved lines. For clear visualization, the original phenotypes reported by individual studies were simplified (Supplementary file 2). Node size is proportional to reported number of genotype–phenotype associations. The biological characteristics of ARDS, including pulmonary inflammation, systemic inflammation, and vascular endothelial activation, are associated with multiple candidate genes for ARDS. Abbreviations: ACE = angiotensin I-converting enzyme; AGER = advanced glycosylation end product-specific receptor; AGT = angiotensin; ANGPT = angiopoietin; ARDS = acute respiratory distress syndrome; CARMIL1 = capping protein regulator and myosin 1 linker 1; CFTR = cystic fibrosis transmembrane conductance regulator; CXCL8 = C-X-C motif chemokine ligand; EPAS1 = endothelial PAS domain protein 1; FER = feline encephalitis virus-related tyrosine kinase; FLT1 = fms-related receptor tyrosine kinase 1; HSPG = heparan sulfate proteoglycan; IL = interleukin; KLK2 = kallikrein-related peptidase 2; SFTP = surfactant protein; LTA = lymphotoxin alpha; MAP3K1 = mitogen-activated protein kinase 1; MBL = mannose binding lectin; NFE2L2 = nuclear factor, erythroid-like, BZIP transcription factor 2; PPFIA = protein tyrosine phosphatase receptor type F interacting protein alpha; SELP = selectin P; THBD = thrombomodulin; TNF = tumor necrosis factor; VCAM1 = vascular cell adhesion molecule 1; VEGFA = vascular endothelial growth factor A; vWF = von Willebrand Factor.
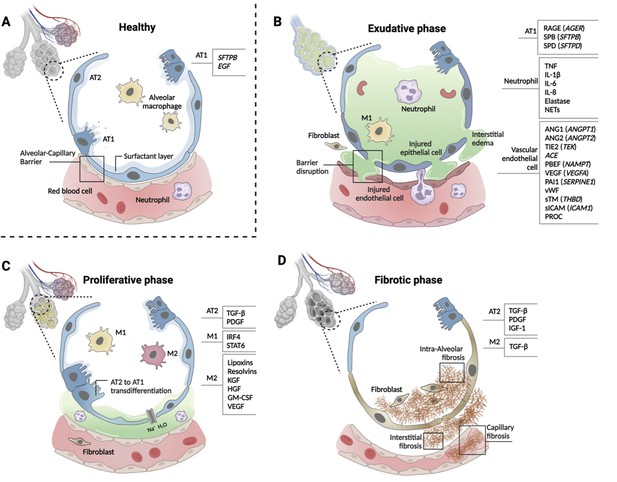
Phases of ARDS pathogenesis.
Panel A shows alveolar–capillary interface in a state of good health. Panels B, C, and D, respectively, show the three phases of ARDS development with candidate biomarkers indicated by cell type. Candidate biomarkers indicate dysregulation following a physiologic insult with transition from (A) health to (B) exudative, (C) proliferative, and (D) fibrotic phases of illness. Patients may present to care in different phases of ARDS development, may progress through phases at different rates, and biology differ depending on cause of ARDS and patient age. Figure was created using Biorender.com. Abbreviations: ACE = angiotensin-converting enzyme; ANG1/ANGPT1 = angiopoietin 1; ANG2/ANGPT2 = angiopoietin 2; ARDS = acute respiratory distress syndrome; AT1 = type I alveolar cell; AT2 = type II alveolar cell; EGF = epidermal growth factor; GM-CSF = granulocyte-macrophage colony-stimulating factor; HGF = hepatocyte growth factor; IGF = insulin like growth factor; IL-1β = interleukin 1 beta; IL-6 = interleukin 6; IL-8 = interleukin 8; IRF4 = interferon regulatory factor 4; KGF = keratinocyte growth factor; M1 = type I macrophage; M2 = type II macrophage; NAMPT = nicotinamide phosphoribosyltransferase; NET = neutrophil extracellular trap; PAI1 = plasminogen activator inhibitor type 1; PBEF = pre-B-cell colony-enhancing factor; PDGF = platelet-derived growth factor; PROC = protein C, inactivator of coagulation factors Va and VIIIa; RAGE = receptor for advanced glycation end-products; SERPINE1 = serpin family E member 1; SFTPB = surfactant protein B; sICAM/ICAM1 = intercellular adhesion molecule 1; SPB = surfactant-associated protein B; SPD = surfactant-associated protein D; STAT6 = signal transducer and activator of transcription 6; sTM = soluble thrombomodulin; TGF-β = transforming growth factor beta; THBD = thrombomodulin; TIE2 (TEK) = tyrosine kinase with immunoglobulin- and EGF-like domains 1; TNF = tumor necrosis factor; VEGF/VEGFA = vascular endothelial growth factor A; vWF = von Willebrand Factor.
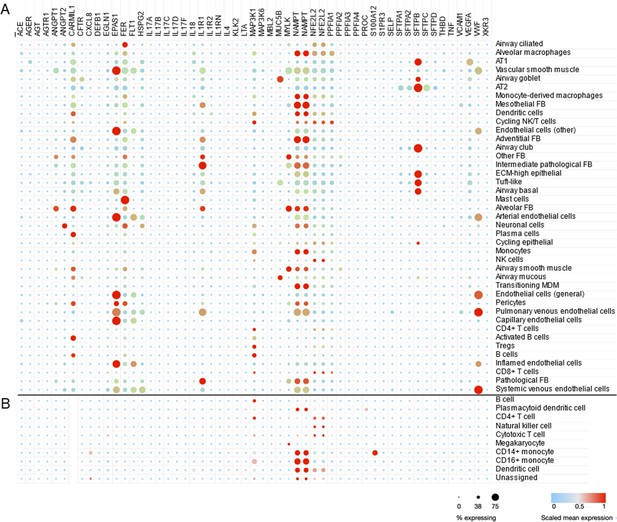
Cell type-specific expression pattern of ARDS candidate genes.
Relative expression levels of ARDS candidate genes are shown across cell types from lung autopsy sample from ARDS caused by COVID-19 (A) and peripheral blood mononuclear cells a generally heathy individual (B). Expression levels are scaled to show relative levels across all cell types from 0 (blue) to 1 (red). The size of circle is proportional to the cells expressing a gene for each cell type (from 0% to 75%). AT1 and AT2: alveolar type I and II cells; FB: fibroblasts; ECM-high: high expression of extracellular matrix components (i.e., COL6A3, COL1A2, and COL3A1); MDM: monocyte-derived macrophages. Source data for this figure are provided in a table, Figure 3—source data 1.
-
Figure 3—source data 1
Source data for Figure 3.
- https://cdn.elifesciences.org/articles/77405/elife-77405-fig3-data1-v1.xlsx
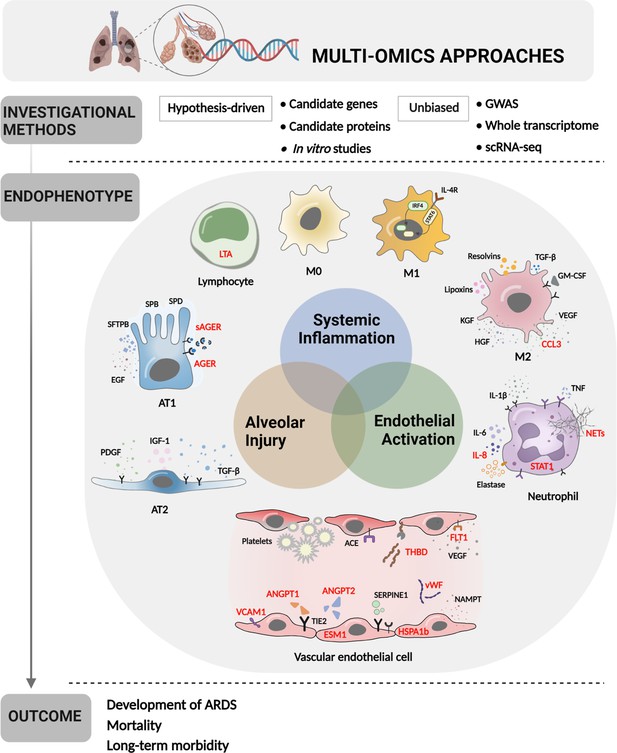
Multi-omics approaches to define endotypes of pediatric acute respiratory distress syndrome (PARDS).
Investigational methods, including hypothesis-driven and unbiased investigations, have led to discovery of multiple contributions to ARDS pathobiology, including systemic inflammation, endothelial activation, and alveolar injury. These processes affect diverse cell and tissue types, including lymphocytes, macrophages, neutrophils, vascular endothelial cells, and two types of alveolar cells. The degree of systemic inflammation, endothelial activation, and alveolar injury across cell and tissue types has helped to define ARDS endotypes, which have been differentially associated with relevant clinical outcomes, including the development of ARDS in patients who have experienced clinical risk factors for the disease, mortality, and long-term morbidity from ARDS. Names of proteins implicated in PARDS as opposed to adult ARDS are depicted in red. Figure was created using Biorender.com. Abbreviations: ACE = angiotensin-converting enzyme; ANGPT1, -2 = angiopoietin 1, 2; CCL3 = C-C motif chemokine ligand 3; ESM1 = endothelial cell-specific molecule 1; FLT1 = fms-related receptor tyrosine kinase 1; GM-CSF = granulocyte-macrophage colony-stimulating factor; HGF = hepatocyte growth factor; HSPA1b = heat shock protein family A (Hsp70) member 1B; IL- = interleukin-; IRF4 = interferon regulatory factor 4; KGF = keratinocyte growth factor; LTA = lymphotoxin alpha; M0, M1, M2 = type 0, I, II macrophage; NAMPT = nicotinamide phosphoribosyltransferase; NET = neutrophil extracellular trap; SERPINE1 = serpin family E member 1; STAT1, -6 = signal transducer and activator of transcription-1,6; TGF-β = transforming growth factor beta; THBD = thrombomodulin; TIE2 = tyrosine kinase with immunoglobulin like and EGF like domains 1; VCAM1 = vascular cell adhesion molecule 1; VEGF = vascular endothelial growth factor A; vWF = von Willebrand Factor.
Tables
Key Pediatric Studies.
| Approach | Candidate Gene(s) | Sample Size | Findings (Adjusted OR and CI) | Author, Journal*, Year |
|---|---|---|---|---|
| HYPOTHESIS-DRIVEN | ||||
| Candidate Gene | TNF, LTA | 490 intubated with sepsis, 610 healthy controls (total 1100) | Protective effect of TNF-308GA against ARDS in infants: OR 0.2, P=0.001 | Azevedo et al., 2012 |
| ACE | 60 with ARDS, 60 healthy controls (total 120, all <15 years) | I/D genotype not increased in ARDS: rate 0.4 (ARDS) vs. 0.3 (controls), P=NS | Cruces et al., 2012 | |
| SFTPB | 395 with pneumonia: 37 requiring mechanical ventilation, 26 with ALI/ARDS | Two linkage disequilibrium-tag SNPs associated with mechanical ventilation: GTGCGCG AOR = 2.62, CI 1–6.8, ATATAAG AOR = 3.1, CI 1–8.9 | Dahmer et al., 2011 | |
| SFTP | 248 children <2 years with acute respiratory failure, 468 newborn healthy controls | 34 interactions among 3 SNPs of SFTPA1, SFTPA2, SFTPC associated with acute respiratory failure (P=0.000000002–0.05) and pulmonary dysfunction after discharge (P=0.00002–0.03) | Gandhi et al., 2020 | |
| ACE | 13 Caucasian children with ARDS, 30 acute hypoxemic respiratory failure, 186 ICU controls (total 216) | I/D polymorphism not associated with acute hypoxemic respiratory failure or ARDS | Plunkett et al., 2008 | |
| Candidate Protein | IL8 | 480 with acute respiratory failure | Increased IL8 associated with mortality, duration mechanical ventilation, ICU LOS but not ARDS diagnosis | Flori et al., 2019 |
| many | 3 cohorts: 46 with sepsis ARDS, 54 with sepsis without ARDS, and 19 ICU controls (total 119) | ANGPT2, ANGPT2/1 ratio higher in ARDS; ANGPT2, ANGPT2/1 ratio, VWF, ESM1 predicted complicated course in sepsis; in sepsis ARDS, FLT1 decreased more quickly and VWF, THBD decreased more slowly in those with complicated course | Whitney et al., 2020a | |
| ANGPT1, ANGPT2, VCAM1, vWF | 2 cohorts of patients with ARDS: 52 direct, 46 indirect lung injury (total 98) | ARDS with indirect lung injury associated with increased ANGPT2/1 ratio, VCAM1, vWF (sensitivity 0.9, CI 0.8–1.0, specificity 0.8, CI 0.7–0.9) | Whitney et al., 2020a | |
| AGER, ANGPT2 | 82 with ARDS | Increased AGER, ANGPT2 associated with non-survival, organ failures in children with ARDS | Yehya et al., 2016 | |
| CCL3, HSPA1b, IL8 | 153 with ARDS | Mortality associated with increased CCL3, HSPA1B, IL8, and older age in children with ARDS | Yehya et al., 2018 | |
| many | 235 with ARDS | Identified MMP profile associated with mortality (AOR 4.0, CI 2.1–7.6) | Zinter et al., 2019 | |
| ANGPT2, VEGF, VWF | 259 with ARDS, 25 status post HCST | Early ANGPT2 (OR 3.7, CI 1.1–11.5) and increasing ANGPT2 associated with mortality (AOR 3.3, CI 1.2–9.2), especially among HCST (AOR 16.3, CI 1.3–198) | Zinter et al., 2016 | |
| In Vitro Studies | Neutrophils and tracheal aspirates from 20 ARDS viral pneumonia with or without bacterial co-infection | In bacterial co-infection: (1) neutrophils more activated with impaired bacterial killing, respiratory burst, (2) aspirates with higher neutrophil elastase and myeloperoxidase, (3) neutrophils transmigrated into aspirate with decreased burst/killing of H. influenzae, S. aureus | Grunwell et al., 2019 | |
| Tracheal aspirates from 42 intubated children with, 35 without ARDS (total 77) | Increased STAT1 phosphorylation, markers of neutrophil degranulation and activation, NET release. Higher airway NETs associated with fever ventilator-free days | Grunwell et al., 2019 | ||
| UNBIASED | ||||
| Gene Expression | 28 intubated with, 26 without ARDS (total 54) | Using tracheal aspirates, a 62-gene signature to identify ARDS was developed to achieve cross-validation AUC 0.8, CI 0.6–0.9 | Grunwell et al., 2021 | |
| 67 with sepsis and acute hypoxemic respiratory failure | Two identified endotypes differentially associated with mortality (OR 8, CI 1.6–41), complicated course (OR 4.2, 1.2–14.9) | Yehya et al., 2019 | ||
| 96 with ARDS | Three identified sub-phentoypes associated with different clinical characteristics, outcomes | Yehya et al., 2020 |
-
Abbreviations: ACE = angiotensin-converting enzyme; AGER = advanced glycosylation end-product specific receptor; ALI = acute lung injury; ANGPT = angiopoietin; AOR = adjusted odds ratio; AUC = area under the receiver operator characteristic curve; ARDS = acute respiratory distress syndrome; CCL3 = C-C motif chemokine ligand 3; CI = 95% confidence interval; ESM1 = endothelial cell specific molecule 1; FLT1 = fms related receptor tyrosine kinase 1; HCST = hematopoietic stem cell transplant; HSPA1B = heat shock protein family A (Hsp70) member 1B; ICU = intensive care unit; IL = interleukin; LOS = length of stay; LTA = lymphotoxin alpha; MMP = matrix metalloproteinase; NET = neutrophil extracellular trap; OR = unadjusted odds ratio; SFTP = surfactant protein; SNP = single nucleotide polymorphism; SFTP = surfactant protein; THBD = thrombomodulin; TNF = tumor necrosis factor; TNFRSF1A = TNF receptor superfamily member 1A; VCAM1 = vascular cell adhesion molecule 1; VWF = von Willebrand Factor.
-
*
Journal Titles are abbreviated according to U.S. National Library of Medicine convention.
Additional files
-
Supplementary file 1
Key adult studies.
- https://cdn.elifesciences.org/articles/77405/elife-77405-supp1-v1.docx
-
Supplementary file 2
Phenotypes associated with ARDS candidate genes from genome-wide association studies including at least 3000 samples.
- https://cdn.elifesciences.org/articles/77405/elife-77405-supp2-v1.docx





