A second DNA binding site on RFC facilitates clamp loading at gapped or nicked DNA
Figures
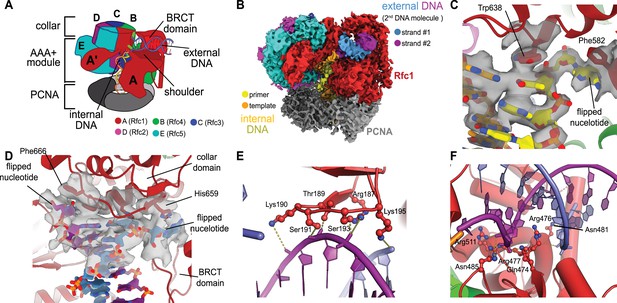
Structure of replication factor C:proliferating cell nuclear antigen (RFC:PCNA) bound to two p/t-DNA molecules.
(A) Schematic of the complex of RFC:PCNA bound to two p/t-DNAs. Melted base pairs are shown as glowing green sticks. (B) Cryo-EM reconstruction of the complex of RFC:PCNA bound to two p/t-DNAs. The strands of the external DNA are shown in slate and purple coloring. (C) The 3′ nucleotide of the primer strand is melted at the internal separation pin. (D) The external DNA binding site also melts DNA. The two melted bases stack against Phe666 and His659. (E) The BRCT domain grips duplex DNA at the external DNA binding site. (F) The ‘shoulder’ region of the AAA+ module grips duplex DNA, with Gln474 and Arg476 inserting into the minor groove, presumably setting the register of DNA.
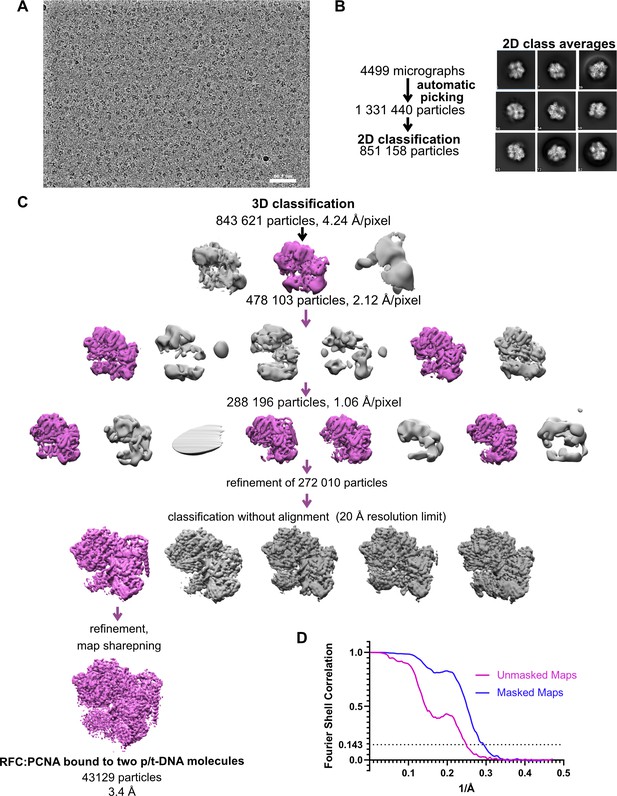
Cryo-EM processing of replication factor C:proliferating cell nuclear antigen (RFC:PCNA) in the presence of p/t DNA.
(A) Downfiltered micrograph taken on a Titan Krios with a Gatan K3 detector. (B) 2D class averages show well-resolved features and different views. (C) Data processing and 3D classification scheme, as described in Gaubitz et al., 2021. Here, a class showing RFC:PCNA bound to two p/t DNA molecules was further refined. The BRCT domain of Rfc1 is most visible in this class. (D) Fourier shell correlation (FSC) curves obtained from postprocessing in Relion for the two halves of the unmasked and masked reconstructions.
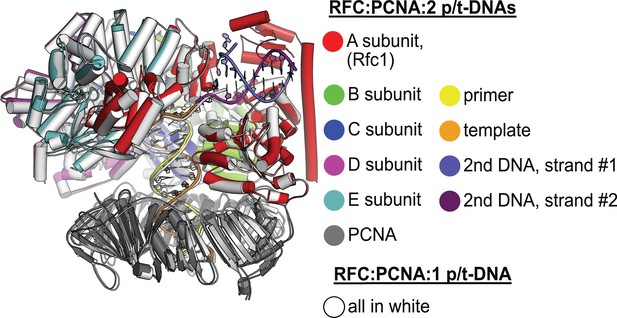
Structural similarity between structures with one or two p/t-DNAs bound.
The structure of replication factor C:proliferating cell nuclear antigen (RFC:PCNA) bound to a single p/t-DNA (white; Gaubitz et al., 2021) compared to the structure reported here bound to two p/t-DNAs (colored). The two structures were superposed only using the AAA+ module of RFC-A; the complex looks largely identical except for the presence of the BRCT domain, linker helix, and second DNA.
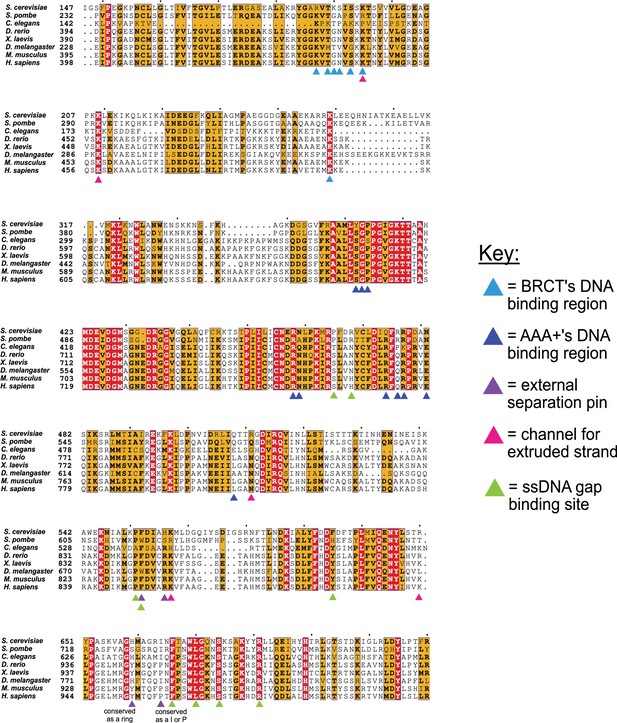
Conservation of key residues for binding DNA.
The key residues in the two DNA binding sites of Rfc1 are conserved among eukaryotes. Sequence alignment shows the conservation of the ‘separation pin’ among eight eukaryotic species. The conserved sequences are marked by blue boxes. The fully conserved residues are in white with a red background, the highly conserved residues are in black with a yellow background, and the less conserved ones are in black. The BRCT’s and AAA+’s DNA binding residues are pointed out by light- and dark-blue arrows, respectively. The external separation pin residues are highlighted by purple arrows. The residues in the channel that interact with the extruded strand are pointed out by pink arrows, and the residues at the ssDNA gap binding site are pointed out by green arrows. Position 659 is conserved as a ring, and Position 664 as I or P.
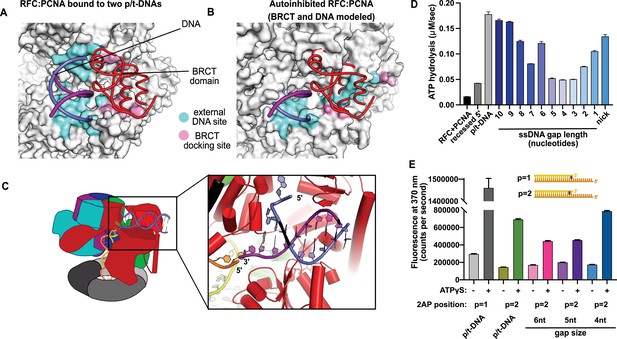
Characterization of replication factor C (RFC) utilization of gapped DNA.
(A, B) The external DNA binding site is incompatible with autoinhibited forms of RFC. (A) The structure of replication factor C:proliferating cell nuclear antigen (RFC:PCNA) bound to two p/t-DNAs, highlighting the external DNA binding site. The residues that interact with DNA and the BRCT domain are shown in light blue and pink, respectively. (B) The external DNA binding site is disabled in the autoinhibited form of RFC. The BRCT domain and DNA duplex were modeled in the same position relative to the AAA+ fold as shown in panel (A). Identical residues of RFC are highlighted in light blue and pink, showing that the binding site is disrupted. Moreover, the collar and A′ regions would sterically clash with the DNA and the BRCT domain. (C) Because the 5′ end of the template strand of the internal DNA is positioned near the 3′ end of a strand in the external site, we hypothesized that the two DNA sites could be connected with a short ssDNA gap of approximately 6 nucleotides. (D) Steady-state ATPase rates of RFC:PCNA in the presence of p/t-DNA, recessed 5′ end DNA, or gapped/nicked DNA. Decreasing gap size results in decreasing ATPase rates until 4 nucleotides of ssDNA, where ATPase rates increase. The trend is smooth and continuous, except for 6-nucleotide single-stranded DNA (ssDNA), which is the size predicted to ideally span the distance between the internal and external sites. (E) 2-Aminopurine (2AP) fluorescence measuring base-flipping in gapped DNA constructs. 2AP at the P = 2 position informs whether base-flipping occurs at P = 1 and/or P = 2 position on the primer strand. 0.5 µM RFC and 2 µM PCNA were incubated with 2AP-containing DNA and fluorescence at 370 nm was recorded. We observe ATP-dependent fluorescence changes, particularly in the 4-nucleotide gapped DNA. Error bars in (D&E) reflect the standard deviation from three replicates.
-
Figure 2—source data 1
ATPase data.
- https://cdn.elifesciences.org/articles/77483/elife-77483-fig2-data1-v2.zip
-
Figure 2—source data 2
2-Aminopurine fluorescence.
- https://cdn.elifesciences.org/articles/77483/elife-77483-fig2-data2-v2.zip
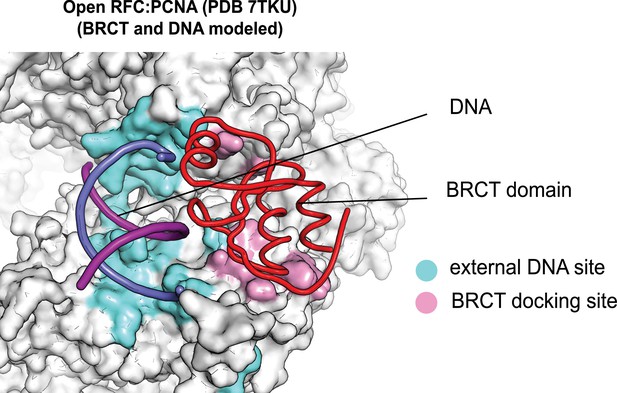
The external DNA binding site is exposed in the open replication factor C:proliferating cell nuclear antigen (RFC:PCNA) structure.
The structure of open RFC:PCNA with no DNA (PDB 7TKU) is overlaid with our structure with two p/t-DNAs bound. The residues that interact with DNA and the BRCT domain (model from two p/t-DNA structure) are shown in light blue and pink, respectively. The shoulder DNA binding site is exposed when RFC:PCNA opens with a crab-claw mechanism.
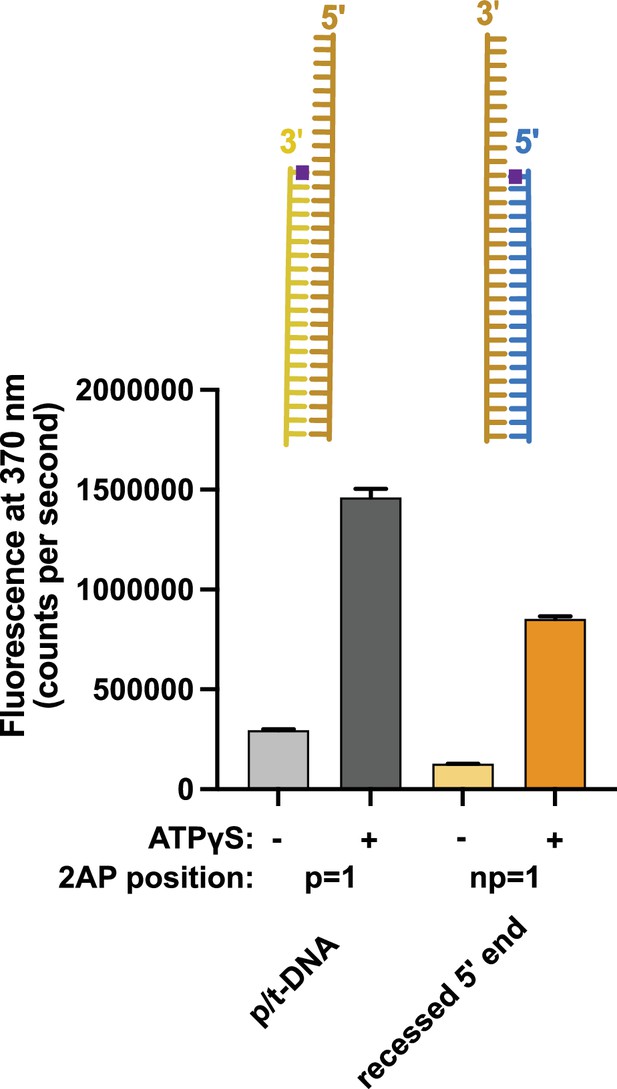
Binding of 5′ recessed ends to replication factor C (RFC).
2-Aminopurine (2AP) fluorescence was measured using DNA with a 3′ recessed end or a 5′ recessed end. The 2AP probe was placed at the 3′ end of the primer or the 5′ end of the template strand, respectively. In the presence of RFC and proliferating cell nuclear antigen (PCNA), we observe an ATP-analog dependent increase in 2AP fluorescence that is comparable to the increase seen with the well-characterized p/t-DNA substrate with 2AP at the 3′ end of the primer strand. This result indicates that 5′ recessed DNA can bind RFC and suggests that the 5′ base is being melted. Because this increase is ATP-dependent and 5′ recessed DNA does not stimulate ATP hydrolysis, this suggests that the melting does not occur at the internal separation pin, but occurs externally in the ATP-dependent BRCT-docked state. Error bars reflect the standard deviation from three replicates.
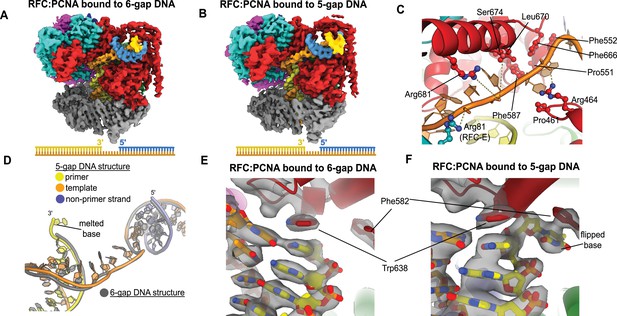
Structures of replication factor C:proliferating cell nuclear antigen (RFC:PCNA) bound to gapped DNA.
(A, B) Cryo-EM reconstruction of the complex of RFC:PCNA bound to DNA with a single-stranded DNA (ssDNA) gap of 6 or 5 nucleotides (6-gap or 5-gap structures). (C) The single-strand gap is specifically bound by residues comprising AAA+, collar, and A′ domains of the A subunit, as well as contacts from the E subunit. (D) DNA conformations in the 5-gap (yellow, orange, and slate) and the 6-gap (gray) structures. The conformations of the DNA are nearly identical except that the 5-gap DNA has melted a single base pair at the internal separation pin so that the ssDNA linker remains 6 nucleotides in length. (E) The 6-gap DNA binds with no melting at the internal separation pin. (F) The internal separation pin melts a single base pair at the internal separation pin.
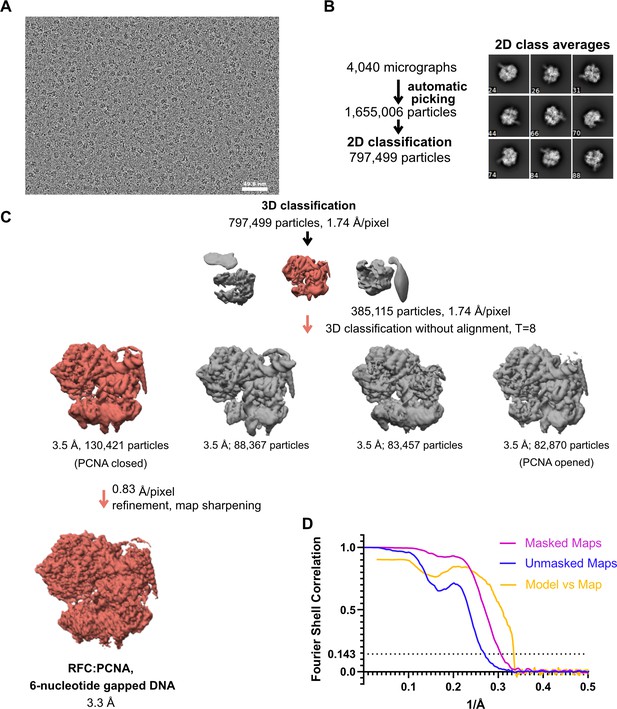
Cryo-EM processing of replication factor C:proliferating cell nuclear antigen (RFC:PCNA) in the presence of double-stranded DNA (dsDNA) with a 6-nucleotide gap.
(A) Micrograph taken on a Talos Arctica with a Gatan K3 detector. (B) 2D class averages show well-resolved features and different side views. (C) The downfiltered reconstruction of RFC.PCNA bound to two p/t DNA molecules was used as a reference for 3D classification. (D) Fourier shell correlation (FSC) curves obtained from postprocessing in Relion for the two halves of the unmasked and masked reconstructions as well as model vs. map curve.
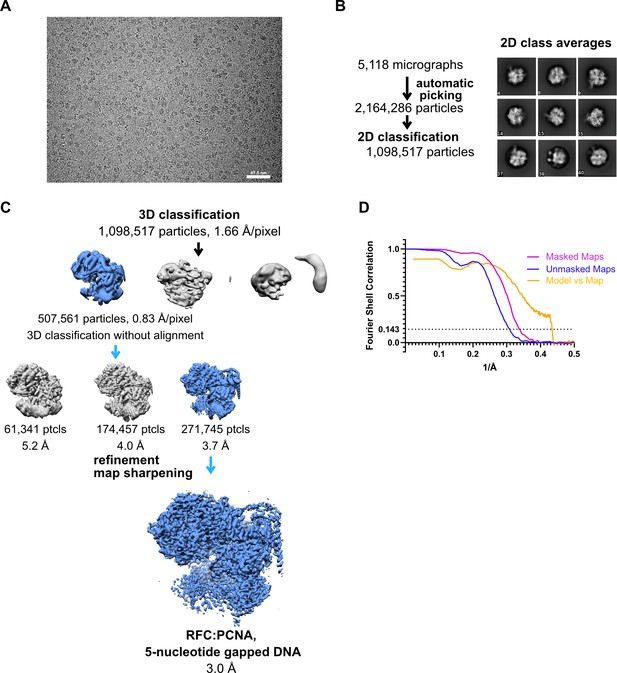
Cryo-EM processing of replication factor C:proliferating cell nuclear antigen (RFC:PCNA) in the presence of double-stranded DNA (dsDNA) with a 5-nucleotide gap.
(A) Micrograph taken on a Titan Krios with a Gatan K3 detector. (B) 2D class averages show different side views. (C) Data processing and 3D classification scheme. The cryo-EM reconstruction of RFC:PCNA bound to dsDNA with a 6-nucleotide gap was used as a 3D reference for 3D classification. (D) Fourier shell correlation (FSC) curves obtained from postprocessing in Relion for the two halves of the unmasked and masked reconstructions, and model vs. map curve.
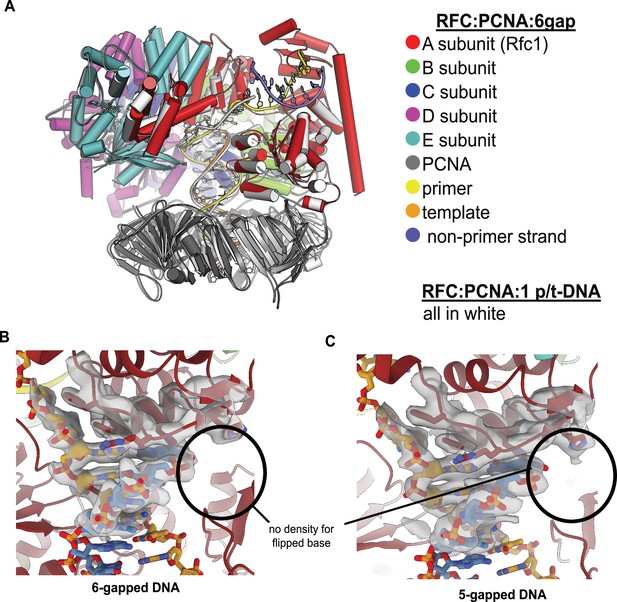
RFC binds 5- or 6-gapped DNA without melting at the external separation pin.
(A) The single-stranded DNA (ssDNA) region in the structure of replication factor C:proliferating cell nuclear antigen (RFC:PCNA) with double-stranded DNA (dsDNA) with a 6-nucleotide gap resembles the template overhang observed in RFC:PCNA bound to a single p/t-DNA. The structure of RFC:PCNA bound to a single p/t-DNA (white; Gaubitz et al., 2021) compared to the structure reported here bound to 6-gapped DNA (colored). The two structures were superposed using the AAA+ module of RFC-A. The ssDNA region of the 6-gapped DNA exits RFC’s central chamber through the A-gate, like the template overhang in the structure of RFC:PCNA bound to one p/t-DNA molecule. (B) Close-up view on the DNA at the external separation pin, cryo-EM density is shown in gray. The 6-gapped DNA binds to RFC:PCNA without melting at the external separation pin (circled area). (C) The 5-gapped DNA also binds to RFC:PCNA without melting at the external separation pin (circled area).
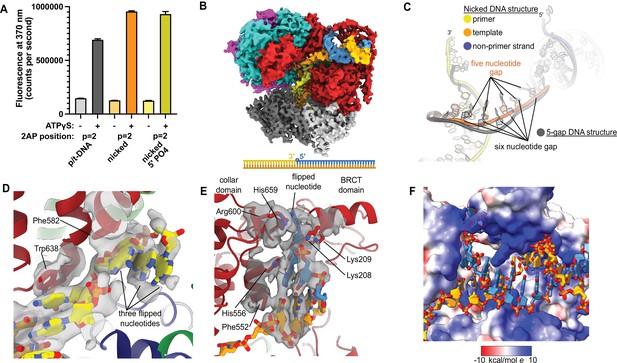
Replication factor C:proliferating cell nuclear antigen (RFC:PCNA) binding to nicked DNA.
(A) RFC melts the primer 3′ end in nicked DNA. 2-Aminopurine (2AP) fluorescence at the primer = 2 position (probe adjacent to the flipped base) showing that the primer strand exhibits even more melting than observed in p/t-DNA. The presence of a 5′ phosphate on the nonprimer strand does not appear to affect base-flipping. Error bars reflect the standard deviation from three replicates. (B) Cryo-EM reconstruction of the complex of RFC:PCNA bound to nicked DNA with a 5′ phosphate on the nonprimer strand. (C) The nicked DNA contains a single-stranded DNA (ssDNA) gap of five nucleotides linking the two duplex binding sites. This is shorter than the structure with 5-gap DNA (gray), which has a 6-nucleotide-long ssDNA linker. (D) At least three base pairs have been melted at the internal separation pin. (E) At least one base pair is melted at the external separation pin. (F) The flipped nucleotide of the nonprimer strand is in a highly electropositive environment to stabilize the backbone.
-
Figure 4—source data 1
2-Aminopurine fluorescence.
- https://cdn.elifesciences.org/articles/77483/elife-77483-fig4-data1-v2.zip
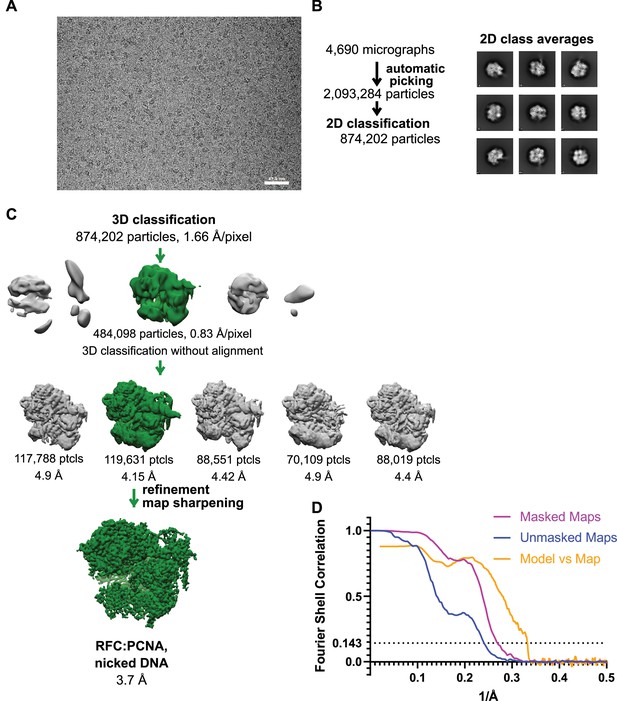
Cryo-EM processing of replication factor C:proliferating cell nuclear antigen (RFC:PCNA) in the presence of nicked double-stranded DNA (dsDNA).
(A) Micrograph taken on a Titan Krios with a Gatan K3 detector. (B) 2D class averages show well-resolved features and different side views. (C) The reconstruction of RFC:PCNA bound to the 5-gapped dsDNA was downfiltered and used as a reference for 3-D classification. (D) Fourier shell correlation (FSC) curves obtained from postprocessing in Relion for the two halves of the unmasked and masked reconstructions as well as the model vs. map curve.
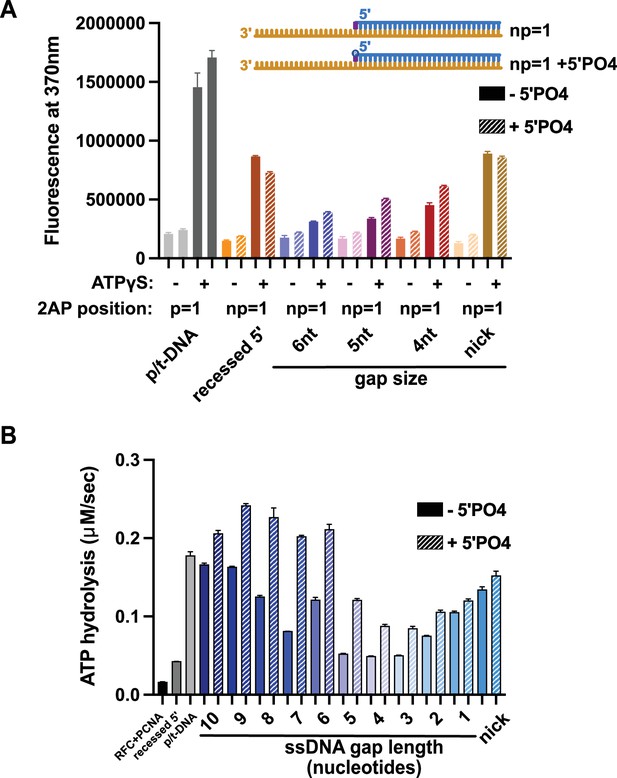
Effect of 5′ phosphate of the nonprimer strand on replication factor C’s (RFC’s) ATPase and DNA-melting activity.
(A) 2-Aminopurine (2AP) on the nonprimer strand with or without a 5′ phosphate (5′PO4) measures base-flipping in gapped or nicked DNA constructs. 2AP at the np = 1 position informs whether base-flipping occurs at the np = 1 position of the nonprimer strand. The solid bars represent DNA without the 5′ phosphate, and the striped bars represent DNA with 5′ phosphate. The RFC- and ATP-dependent fluorescence of DNA with a 5′ phosphate on the nonprimer strand compared to DNA without the matching 5′ phosphate remains similar. (B) Steady-state ATPase rates of RFC:PCNA in the presence of p/t-DNA, recessed 5′, or nicked/gapped DNA constructs with or without a 5′ phosphate on the nonprimer strand. The 5′ phosphate slightly increased ATPase activity. Error bars in (A&B) reflect the standard deviation from three replicates.
-
Figure 4—figure supplement 2—source data 1
2-Aminopurine fluorescence.
- https://cdn.elifesciences.org/articles/77483/elife-77483-fig4-figsupp2-data1-v2.zip
-
Figure 4—figure supplement 2—source data 2
ATPase data.
- https://cdn.elifesciences.org/articles/77483/elife-77483-fig4-figsupp2-data2-v2.zip
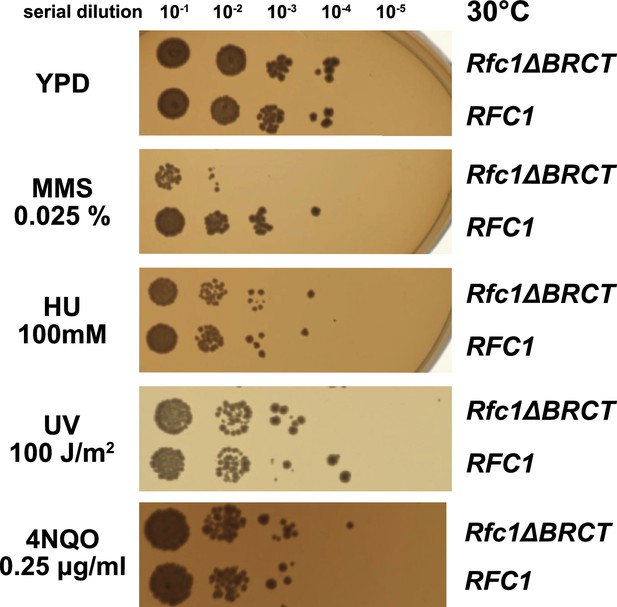
Deletion of the BRCT domain of Rfc1 results in a DNA damage repair defect.
Yeast carrying the sole copy of the RFC1 gene on a plasmid were subjected to various treatments that stress DNA metabolism. Serial 10-fold dilutions of yeast cultures with a starting OD of 0.2 were spotted onto YPD plates with or without additives and grown for 3 days. Rfc1-∆BRCT yeast exhibit a growth defect with the DNA alkylating agent methyl methanesulfonate (MMS), but not with hydroxyurea (HU), ultraviolet radiation (UV), or 4-nitroquinoline 1-oxide (4NQO). Various other conditions are shown in Figure 5—figure supplement 1.
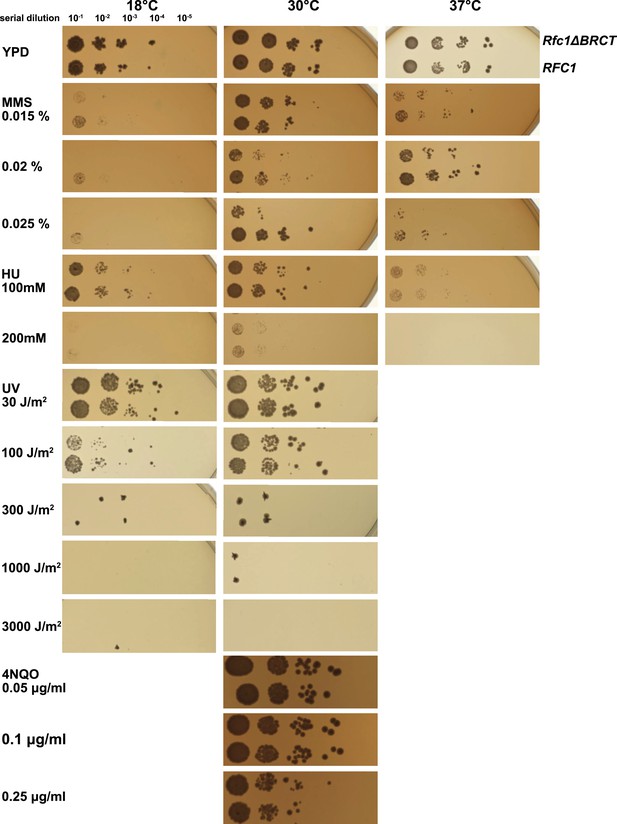
Deletion of the BRCT domain of Rfc1 results in a DNA damage repair defect.
Yeast carrying the sole copy of the RFC1 gene on a plasmid were subjected to various treatments that stress DNA metabolism. Multiple concentrations of the DNA damaging agents were used at different temperatures, including 0.015, 0.02, and 0.025% methyl methanesulfonate (MMS), 100 and 200 mM hydroxyurea (HU), at 18, 30, and 37°C, 30, 100, 300, 1000, and 3000 J/m2 ultraviolet radiation (UV) at 18 and 30°C, and 0.05, 0.1, and 0.25 μg/ml 4-nitroquinoline 1-oxide (4NQO) at 30°C. Rfc1-∆BRCT yeast exhibit a growth defect only with the DNA alkylating agent MMS, but not with HU, UV, or 4NQO. The MMS phenotype is not dependent on temperature.
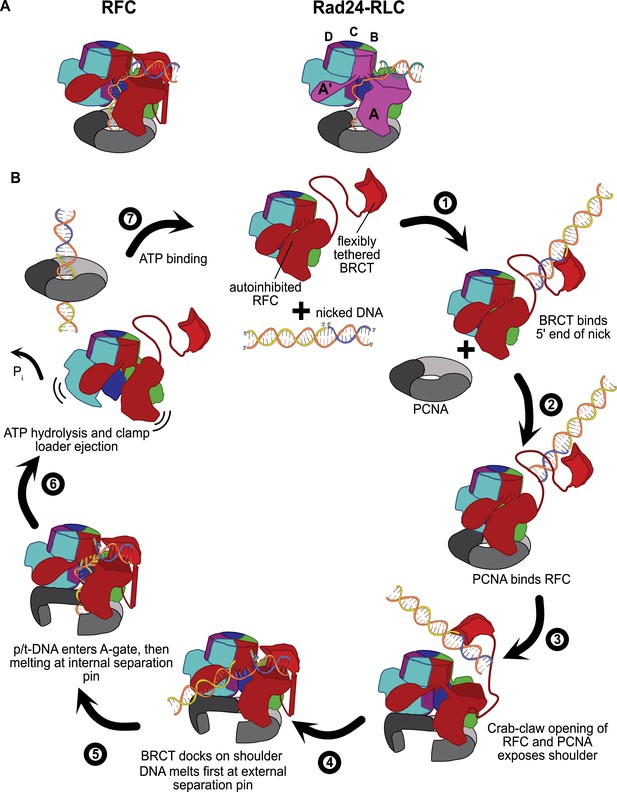
Insights into clamp loader evolution and mechanism from the discovery of the external DNA binding site.
(A) The external binding site of replication factor C (RFC) is similar to that recently reported for Rad24-RLC (Zheng et al., 2021; Castaneda et al., 2021), suggesting that a straightforward mechanism for the evolution of Rad24-RLC’s ability to load clamps at 5′-recessed DNA. (B) A speculative model for proliferating cell nuclear antigen (PCNA) loading by RFC onto nicked DNA. Melted bases are shown in glowing green. The loosely tethered BRCT domain can capture a DNA segment and keep it in close proximity. Once RFC binds and opens PCNA, the external DNA binding site is formed. DNA melting at the external and internal separation pins allows for nicked DNA to fully engage RFC, thereby activating ATP hydrolysis, clamp closure, and RFC release. See ‘Discussion’ for a more detailed description.
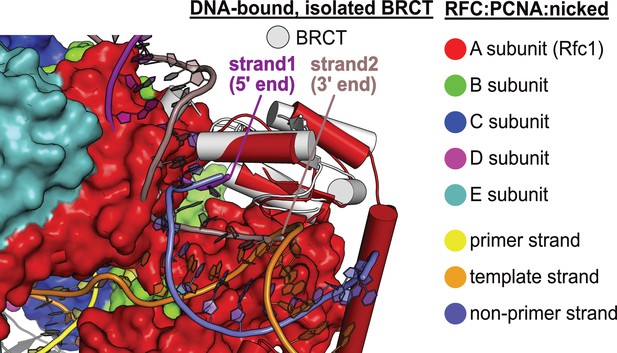
DNA binding mode of free BRCT is incompatible with the conformation of open replication factor C (RFC).
The structure of RFC1’s free BRCT domain (PDB code: 2K7F, Kobayashi et al., 2010) bound to DNA compared to the structure of replication factor C:proliferating cell nuclear antigen (RFC:PCNA) bound to nicked DNA. The structures were superposed on the BRCT domain. The DNA binding mode of free BRCT is incompatible with the conformation of RFC reported here. The DNA clashes with the collar domain of A subunit.
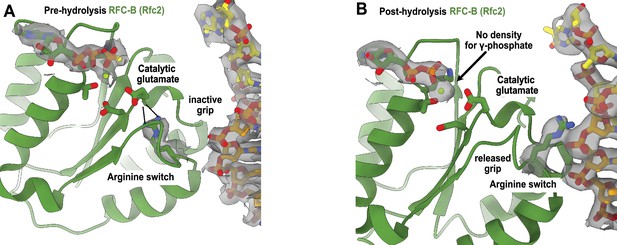
Arginine switch residue is flipped into the active conformation.
(A) The active site of RFC-B is in pre-hydrolysis state in the structure of replication factor C:proliferating cell nuclear antigen (RFC:PCNA) bound to p/t-DNA, and the conserved arginine switch residue holds the catalytic glutamate in an inactive conformation (PDB ID: 7TID). (B) Here, 5-gapped DNA is bound and the arginine switch instead interacts with DNA, releasing its grip from the catalytic glutamate. The catalytic glutamate is now considered to be in an active conformation. Accordingly, there is no density for the γ-phosphate of ATPγS in this conformation, indicating that hydrolysis has occurred at this site. We do not observe this hydrolysis in the structures bound to two p/t-DNAs or to nicked DNA, indicating that the presence of DNA at the external binding site is not driving ATP hydrolysis.
Tables
Cryo-EM data collection, processing, and model statistics.
| Dataset | RFC:PCNA with p/t DNA | RFC:PCNA with dsDNA with a 6-nucleotide gap | RFC:PCNA with dsDNA with a 5-nucleotide gap | RFC:PCNA with nicked DNA |
|---|---|---|---|---|
| Magnification | 81,000 | 45,000 | 105,000 | 105,000 |
| Voltage (keV) | 300 | 200 | 300 | 300 |
| Cumulative exposure (e–/Å2) | 40 | 45 | 48 | 49 |
| Detector | K3 | K3 | K3 | K3 |
| Pixel size (Å) | 1.06 | 0.87 | 0.83 | 0.83 |
| Defocus range (μm) | –1.2 to –2.3 | –1–2.2 | –1–2.2 | –1–2 |
| Micrographs used (no.) | 4499 | 4040 | 5118 | 4690 |
| Initial particle images (no.) | 1,331,440 | 797,499 | 1,098,517 | 874,202 |
| Symmetry | C1 | |||
| Class name | RFC:PCNA bound to two p/t DNA molecules | RFC:PCNA bound to dsDNA with a 6-nucleotide gap | RFC:PCNA bound to dsDNA with a 5-nucleotide gap | RFC:PCNA bound to nicked DNA |
| Final refined particles (no.) | 43,129 | 130,421 | 271,745 | 119,631 |
| Map resolution (Å, FSC 0.143) | 3.4 | 3.3 | 3.0 | 3.7 |
| Model-Map CC_mask | - | 0.83 | 0.82 | 0.8 |
| Bond lengths (Å), angles (°) | - | 0.003, 0.619 | 0.003, 0.596 | 0.003, 0.662 |
| Ramachandran outliers, allowed, favored | - | 0.0, 2.05, 97.95 | 0.0, 1.53, 98.47 | 0.0, 1.79, 98.21 |
| Poor rotamers (%), MolProbity score, Clashscore (all atoms) | - | 0.04, 1.61, 12.23 | 0.04, 1.55, 10.67 | 0.33, 1.60, 12.10 |
| EMDB ID | EMD-26280 | EMD-26298 | EMD-26302 | EMD-26297 |
| PDB ID | - | 7U1A | 7U1P | 7U19 |
-
RFC, replication factor C; PCNA, proliferating cell nuclear antigen; dsDNA, double-stranded DNA; FSC, Fourier shell correlation.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | BL21(DE3) | Novagen | 69450 | Chemically competent cells |
| Recombinant DNA reagent | pET(11a)-RFC[2+3+4] (plasmid) | Finkelstein et al., 2003 | Expression plasmid | |
| Recombinant DNA reagent | pLANT-2/RIL[1+5] (plasmid) | Finkelstein et al., 2003 | Expression plasmid | |
| Recombinant DNA reagent | pRS413-RFC1 (plasmid) | Gaubitz et al., 2021 | Plasmid for yeast expression of Rfc1 from endogenous promoter | |
| Recombinant DNA reagent | pRS413-RFC1RFC1-∆BRCT (plasmid) | This study | Plasmid for yeast expression of Rfc1 from endogenous promoter | |
| Strain, strain background (Saccharomyces cerevisiae) | BY4743 his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 LYS2/lys2Δ0 met15Δ0/MET15 ura3Δ0/ura3Δ0 ∆rfc1::KanMX4/ RFC1 (YOR217W) | Dharmacon | YSC1055 (22473) | Yeast Heterozygous Collection |
| Software, algorithm | RELION | doi: 10.7554/eLife.42166 | Relion 3.1 | |
| Software, algorithm | cisTEM | doi: 10.7554/eLife.35383 | cisTEM-1.0.0-beta | https://cistem.org/software |
| Software, algorithm | Ctffind | doi: 10.1016j.jsb.2015.08.008 | Ctffind 4.1 | |
| Software, algorithm | UCSF Chimera | UCSF, doi: 10.1002/jcc.20084 | http://plato.cgl.ucsf.edu/chimera/ | |
| Software, algorithm | ChimeraX | UCSF, doi: 10.1002/pro.3943 | ChimeraX-1.2 | https://www.cgl.ucsf.edu/chimerax/ |
| Software, algorithm | COOT | doi:10.1107/S0907444910007493 | Coot-0.9.4 | http://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| Software, algorithm | Phenix | doi:10.1107/S0907444909052925 | Phenix-1.20.1–4487 | https://phenix-online.org |
| Software, algorithm | PyMOL | PyMOL Molecular Graphics System, Schrodinger LLC | https://www.pymol.org/ | |
| Software, algorithm | GraphPad Prism | GraphPad | GraphPad Prism 9.2.1 | http://www.graphpad.com/ |
| Peptide, recombinant protein | Pyruvate kinase | Calzyme | 107A0250 | Enzyme used in ATPase assay |
| Peptide, recombinant protein | Lactate dehydrogenase | Worthington Biochemical Cooperation | LS002755 | Enzyme used in ATPase assay |
| Other | Phosphoenol-pyruvic acid monopotassium salt | Alfa Aesar | B20358 | Reagent used in ATPase assay |
| Chemical compound, drug | Methyl methanesulfonate (MMS) | Sigma-Aldrich | 66-27-3 | https://www.sigmaaldrich.com/US/en/product/aldrich/129925 |
| Chemical compound, drug | Hydroxyurea (HU) | Sigma-Aldrich | 127-07-1 | https://www.sigmaaldrich.com/US/en/product/sigma/h8627 |
| Chemical compound, drug | 4-Nitroquinoline (4NQO) | Fisher Scientific | AC203790010 | https://www.fishersci.com/shop/products/4-nitroquinoline-n-oxide-98-thermo-scientific-3/AC203790010 |
DNA sequences.
| Template | Sequence | Primer | Sequence | Nonprimer | Sequence | Name in assay |
|---|---|---|---|---|---|---|
| Template30, T30 | TTTTTTTTTTTATGTA CTCGTAGTGTCTGC | Primer20-2AP-0 | GCAGACACTACG AGTACAT/32AmPu/ | p/t-DNA P = 1 | ||
| Template30-T-1 | TTTTTTTTTTTTTGTA CTCGTAGTGTCTGC-3’ | Primer20-2AP-1 | GCAGACACTACG AGTACA/i2AmPr/A | p/t-DNA P = 2 | ||
| Template50-ap_gapped2 | TTGTGGGTAGAT AAATACAGACCTAA GTCCTTTGTA CTCGTAGTGTCTGC | Primer20-2AP-1 | GCAGACACTACG AGTACA/i2AmPr/A | 3'PrimerB24_gapped | AGGTCTGTATTT ATCTACCCACAA | 6 nt gap P = 2 |
| Same as above | Same as above | 3'PrimerB25_gapped | TAGGTCTGTATTT ATCTACCCACAA | 5 nt gap P = 2 | ||
| Same as above | Same as above | 3'PrimerB26_gapped | TTAGGTCTGTATTT ATCTACCCACAA | 4 nt gap P = 2 | ||
| Same as above | Same as above | 3'PrimerB30_gapped | GGACTTAGGTCTGTA TTTATCTACCCACAA | Nicked P = 2 | ||
| Same as above | Same as above | 3'PrimerB30_gapped_P | /5Phos/GGACTTA GGTCTGTA TTTATCTACCCACAA | Nicked 5’ PO4 P = 2 | ||
| Template30-3'-T | TATGTACTCGTAGTG TCTGTTTTTTTTTTT | Primer20-2AP-20 | /52AmPr/CAGACA CTACGAGTACATA | Recessed 5' P = 1 | ||
| Template50-ap_nick_np1 | TTGTGGGTAGATA AATACAGACC TAAGTCTTATGTAC TCGTAGTGTCTGC | Primer20-2AP-0 | GCAGACACTACG AGTACAT/32AmPu/ | p/t-DNA P = 1 (design 2, used for 2AP + PO4) | ||
| 3'PrimerB30_np1 | AGACTTAGGTCTGTA TTTATCTACCCACAA | Recessed 5’ np = 1 | ||||
| Template50-ap_nick_np1 | TTGTGGGTAGATAA ATACAGACC TAAGTCTTATGTACT CGTAGTGTCTGC | Primer20-2AP-0 | GCAGACACTACG AGTACAT/32AmPu/ | 3'PrimerB30_np1 | AGACTTAGGTCTGTA TTTATCTACCCACAA | Nicked np = 1 |
| Template50-ap_6gap_np1 | TTGTGGGTAGATA AATACAGACCT AAGTCTTTTTTTTAT GTACTCGTAGTGTCTGC | Same as above | Same as above | 6 nt gap np = 1 | ||
| Template50-ap_5gap_np1 | TTGTGGGTAGATA AATACAGACCTAAG TCTTTTTTTATGTA CTCGTAGTGTCTGC | Same as above | Same as above | 5 nt gap np = 1 | ||
| Template50-ap_4gap_np1 | TTGTGGGTAGATA AATACAGACCTAA GTCTTTTTTATGTA CTCGTAGTGTCTGC | Same as above | Same as above | 4 nt gap np = 1 | ||
| 3'PrimerB30_2AP_np1_P | /5Phos//i2AmPr/GAC TTAGGTCTGTA TTTATCTACCCACAA | Nonprimer np = 1 + PO4 (2AP) | ||||
| Template50_gapped | TTGTGGGTAGATAA ATACAGACCTAA GTCCTTGAATGCC GCGTGCGTCCC | 5’Primer 20_gapped | GGGACGCACGC GGCATTCAA | p/t-DNA (used in ATPase assay) | ||
| Same as above | Same as above | 3'PrimerB20_gapped | CTGTATTTATCTACCCACAA | 10 gap | ||
| Same as above | 5'Primer 21_gapped | GGGACGCACGC GGCATTCAAG | Same as above | 9 gap | ||
| Same as above | 5'Primer22_gapped | GGGACGCACGCG GCATTCAAGG | Same as above | 8 gap | ||
| Same as above | 5'Primer23_gapped | GGGACGCACGCG GCATTCAAGGA | Same as above | 7 gap | ||
| Same as above | 5’Primer24_gapped | GGGACGCACGC GGCATTCAAGGAC | Same as above | 6 gap (ATPase, cryo-EM) | ||
| Same as above | 5’Primer25_gapped | GGGACGCACGC GGCATTCAAGGACT | Same as above | 5 gap (ATPase, cryo-EM) | ||
| Same as above | 5'Primer26_gapped | GGGACGCACGCGGC ATTCAAGGACTT | Same as above | 4 gap | ||
| Same as above | 5'Primer27_gapped | GGGACGCACGCGGC ATTCAAGGACTTA | Same as above | 3 gap | ||
| Same as above | 5'Primer28_gapped | GGGACGCACGCGGC ATTCAAGGACTTAG | Same as above | 2 gap | ||
| Same as above | 5'Primer29_gapped | GGGACGCACGCGGC ATTCAAGGACTTAGG | Same as above | 1 gap | ||
| Same as above | 5'Primer30_gapped | GGGACGCACGCGGC ATTCAAGGACTTAGGT | Same as above | Nicked | ||
| 3'PrimerB20_gapped_P | /5Phos/CTGTATTTA TCTACCCACAA | Nonprimer with 5’ phosphate (ATPase) | ||||
| Primer20-3'-T-10ext | GCAGACACTACGAG TACATTTTTTTTTTTT | Template20-5'-A | AATGTACTCGT AGTGTCTGC | Recessed 5’ | ||
| Template50-ap_gapped2 | TTGTGGGTAGATAA ATACAGACCTAAG TCCTTTGTACTCG TAGTGTCTGC | Primer20-1 | GCAGACACTA CGAGTACAAA | 3'PrimerB30_gapped_P | /5Phos/GGACTTAGGTCTGT ATTTATCTACCCACAA | Nicked 5’ PO4 (cryo-EM) |





