Lack of Tgfbr1 and Acvr1b synergistically stimulates myofibre hypertrophy and accelerates muscle regeneration
Figures
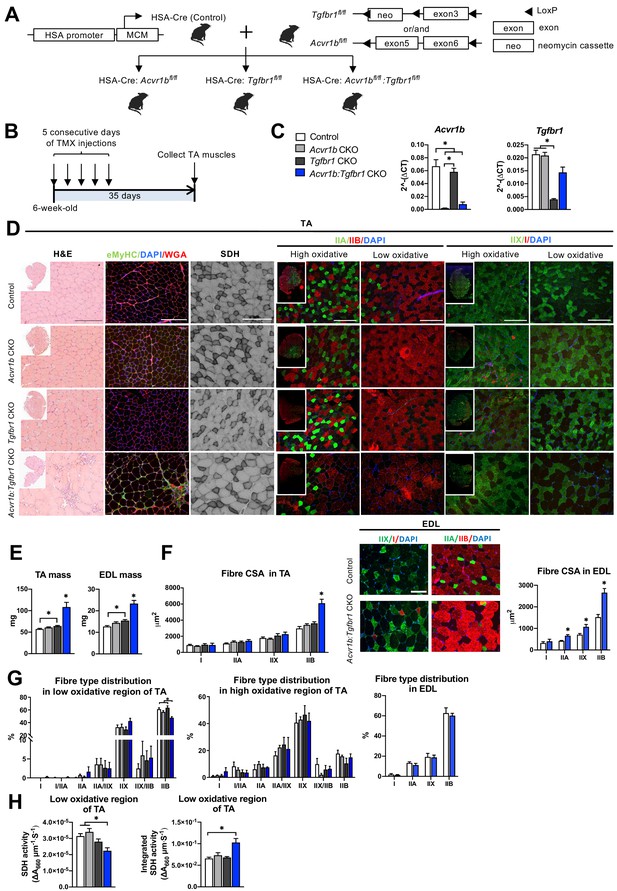
Simultaneous knockout of both Acvr1b and Tgfbr1 caused muscle hypertrophy.
(A) Scheme showing cross-breeding of HSA-Cre mouse line with conditional knockout mouse lines Acvr1bfl/fl and Tgfbr1fl/fl. LoxP sites are indicated by black arrows. A loxP-flanked neomycin (neo) cassette is inserted upstream of exon3 of Acvr1b genome. (B) Scheme demonstrating receptor knockout induced by tamoxifen (TMX) injection for consecutive 5 days. (C) Relative mRNA expression of Acvr1b and Tgfbr1 in TA muscles of experimental groups. (D) Histology stainings of TA muscles 35 days after first TMX injection. H&E staining and immunofluorescent staining of eMyHC (green) of TA showed regenerative regions containing eMyHC+ myofibres with central nuclei (DAPI, blue) in Acvr1b:Tgfbr1 CKO mice, wheat glucose agglutinin (WGA, red) was used to visualise cell membranes and ECM. Acvr1b:Tgfbr1 CKO mice showed lower staining intensity for SDH activity in low oxidative region of TA. MyHCs staining demonstrated type IIA (green), IIB (red), IIX (green) and I (red) myofibres in low and high oxidative regions of TA. Scale bars = 250 μm. (E) TA and EDL muscle mass and myofibre cross-sectional areas (CSAs) were increased in Acvr1b:Tgfbr1 CKO mice. (F) In TA, specifically CSA of type IIB myofibres was increased in Acvr1b:Tgfbr1 CKO animals, while in EDL CSA of all type II myofibres was increased. Myofibre types were stained in EDL. (G) Percentage of type IIB in low oxidative region of TA was reduced. No differences were observed in myofibre distribution in high oxidative region of TA or EDL. (H) SDH activity (absorbance units (∆A660) per micrometer section thickness per second of incubation time (∆A660∙μm–1∙s–1)) was decreased, while the integrated SDH activity, SDH activity multiplied by CSA (∆A660∙μm∙s–1), increased in low oxidative region of TA of Acvr1b:Tgfbr1 CKO animals. N = 5–8 mice. Results are presented as mean + SEM. *: p < 0.05. Significant difference between individual groups is indicated by lines with a *. Single * indicates significant difference compared to all other groups.
-
Figure 1—source data 1
Quantification of Acvr1b and Tgfbr1 gene expression levels in TA and myofiber phenotype in TA and EDL in absence of injury.
- https://cdn.elifesciences.org/articles/77610/elife-77610-fig1-data1-v2.xlsx
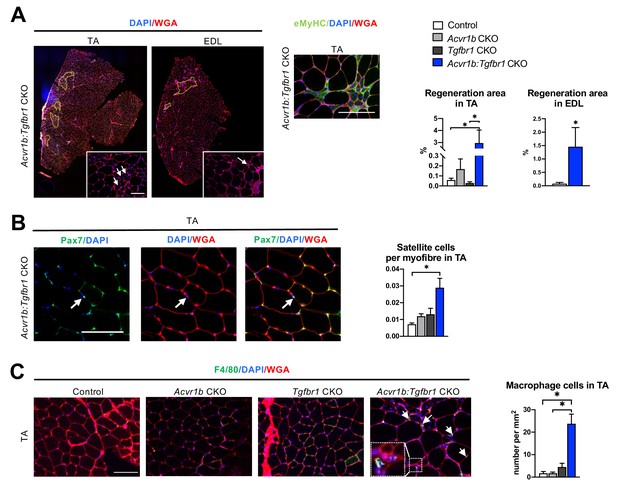
Increased heterogeneity of cell types was found in both TA and EDL of Acvr1b:Tgfbr1 CKO animals.
(A) Regions with spontaneously regenerating myofibres (circled by yellow dash lines) with central nuclei (indicated by arrows) were particularly present in low oxidative region of TA and EDL of Acvr1b:Tgfbr1 CKO animals. (B) Increased number of Pax7+ cells per myofibre was found in TA of Acvr1b:Tgfbr1 CKO mice. (C) IF staining of F4/80 (green) showed an increased number of macrophages (indicated by arrows) in TA muscle per mm2 CSA of Acvr1b:Tgfbr1 CKO mice compared to control. Macrophages (image with higher magnification on the left corner) were mainly located around myofibres with central nuclei. Scale bar = 100 µm. N = 5–8 mice. Results are presented as mean + SEM. *. p < 0.05. Significant differences between individual groups are indicated by lines with a *. Single * indicates significant difference compared to all other groups at the same time point.
-
Figure 2—source data 1
Quantification of number of regenerating myofibres, satellite cells and macrophages in absence of injury.
- https://cdn.elifesciences.org/articles/77610/elife-77610-fig2-data1-v2.xlsx
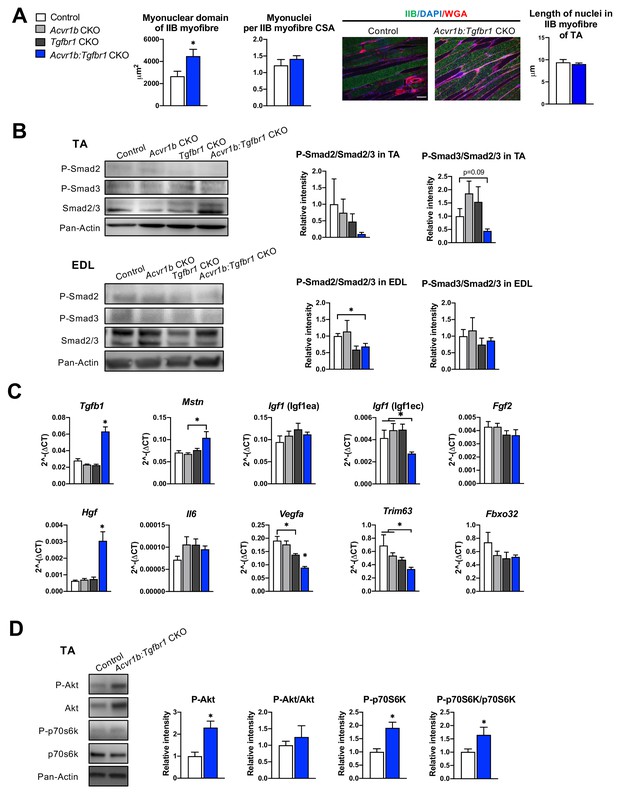
Effects of simultaneous knockout of both Acvr1b and Tgfbr1 on myonuclear number and signalling for protein synthesis as well as degradation.
(A) No differences in myonuclear lengths were observed in longitudinal sections of TA type IIB myofibres of Acvr1b:Tgfbr1 CKO compared to control animals. This indicates that simultaneous knockout of Acvr1b:Tgfbr1 CKO did not affect the number of myonuclei per myofibre and that the myonuclear domain (i.e. cross-sectional area/ nuclei (μm2)) was almost doubled. Scale bar = 100 μm. (B) Western blot analysis for Smad2/3 phosphorylation in TA and EDL muscle. (C) Relative gene expression of growth factors in non-injured muscle. (D) Western blot analysis of phosphorylated and total Akt and p70S6K in TA muscles. Results are presented as mean + SEM. N = 5–8 mice. *: p < 0.05. Significant differences between individual groups are indicated by lines with a *. Single * indicates significant difference compared to all other groups at the same time point.
-
Figure 3—source data 1
Quantification of myonuclear domain, qPCR of growth factors and Western blot of P-Smad2/3, Smad2/3, P-Akt, Akt, P-p70s6k, and p70s6k in absence of injury.
- https://cdn.elifesciences.org/articles/77610/elife-77610-fig3-data1-v2.xlsx
-
Figure 3—source data 2
Raw Western blot image of (A) P-Smad2, (C) P-Smad3 and (E) Smad2/3, (G) pan-Actin.
Labelled images of (B) P-Smad2, (D) P-Smad3 and (F) Smad2/3 and (H) pan-Actin of tibialis anterior (TA) muscle in groups of control (con), Acvr1b CKO (A), Tgfbr1 CKO (T) and Acvr1b: Tgfbr1 CKO (dKO).
- https://cdn.elifesciences.org/articles/77610/elife-77610-fig3-data2-v2.pdf
-
Figure 3—source data 3
Raw Western blot image of (A) P-Smad2, (C) P-Smad3, and (E) Smad2/3, (G) pan-Actin.
Labelled images of (B) P-Smad2, (D) P-Smad3, and (F) Smad2/3 and (H) pan-Actin of extensor digitorum longus muscle (EDL) in groups of control (con), Acvr1b CKO (A), Tgfbr1 CKO (T), Acvr1b: Tgfbr1 CKO (dKO) and positive control sample (pos).
- https://cdn.elifesciences.org/articles/77610/elife-77610-fig3-data3-v2.pdf
-
Figure 3—source data 4
Raw Western Blot image of (A) P-AKT, (C) AKT and (E) P-p70s6k, (G) p70s6k and (I) pan-Actin.
Labelled images of (B) P-AKT, (D) AKT and (F) P-p70s6k, (H) p70s6k and (I) pan-Actin of TA in groups of control (con), Acvr1b: Tgfbr1 CKO (dKO) and positive control sample (pos).
- https://cdn.elifesciences.org/articles/77610/elife-77610-fig3-data4-v2.pdf
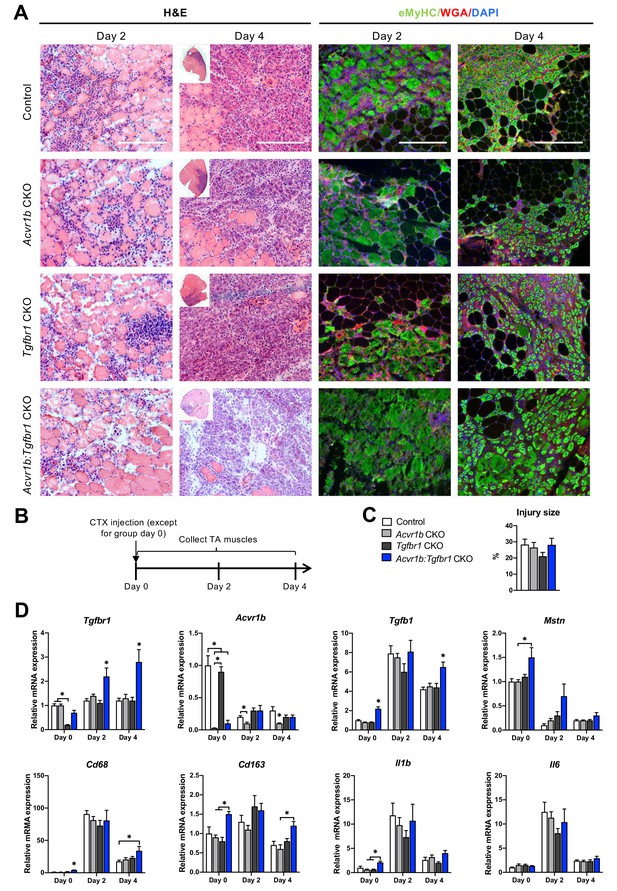
Immune response was slightly enhanced in muscle of Acvr1b:Tgfbr1 CKO mice.
(A) Representative images of H&E and eMyHC staining of TA sections at 2 and 4 days after CTX injection. Scale bars = 250 μm. (B) Scheme shows CTX injection in TA and sample collection. (C) Percentage of injury area was not significantly different between groups. (D) Relative gene expressions in TA in the absence (day 0) or presence of CTX injection after 2 and 4 days. Results are presented as mean + SEM. N = 5–8 mice, *: p < 0.05. Significant differences between individual groups are indicated by lines with a *. Single * indicates significant difference compared to all other groups at the same time point.
-
Figure 4—source data 1
Quantification of injury size and qPCR for myogenic genes in TA at day 0,2 and 4.
- https://cdn.elifesciences.org/articles/77610/elife-77610-fig4-data1-v2.xlsx
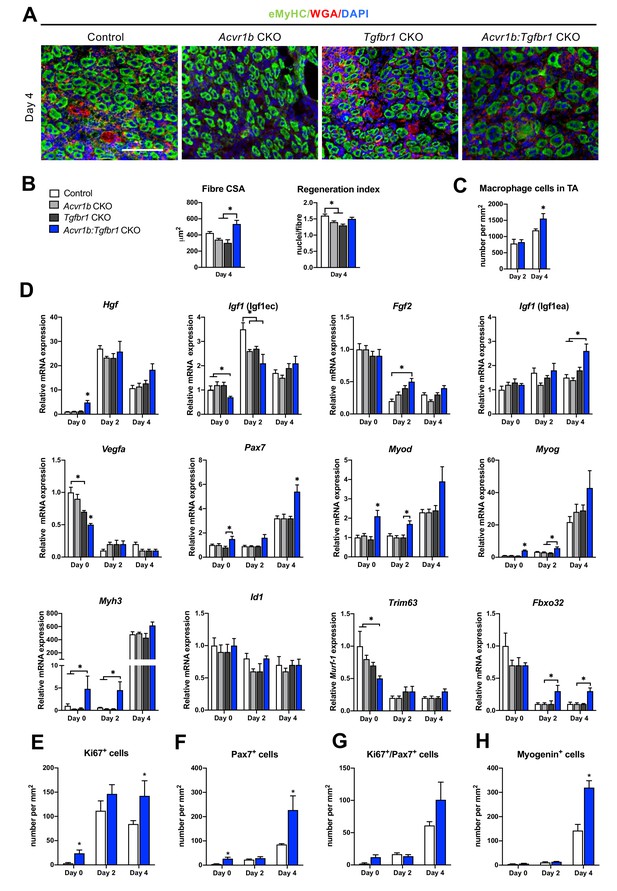
Acvr1b:Tgfbr1 CKO mice showed enhanced CSA of regenerating myofibres and early enhanced expression of myogenic genes and differentiating cells after acute injury.
(A) IF staining images represent eMyHC+ myofibres 4 days after CTX injection. Scale bar = 100 μm. (B) CSA of eMyHC+ myofibres in injured area increased in Acvr1b:Tgfbr1 CKO mice compared to Acvr1b CKO and Tgfbr1 CKO animals, while RI was decreased in both Acvr1b CKO and Tgfbr1 CKO mice compared to controls. (C) Number of macrophages was quantified in the injured area. (D) Relative gene expression in TA in absence (day 0) or presence of CTX injection after 2 and 4 days are presented. Increased number of Ki67+ cells (E) and Pax7+ (F) cells were found in TA of Acvr1b:Tgfbr1 CKO mice in absence of injury as well as 4 days after CTX injection. (G) Four days post injury, number of Ki67+/Pax7+ cells was not different between control and Acvr1b:Tgfbr1 CKO mice. (H) More Myogenin+ cells were found in injured area of Acvr1b:Tgfbr1 CKO mice on day 4 post injury. Results are presented as mean + SEM. N = 5–8 mice, *: p < 0.05. Significant differences between individual groups are indicated by lines with a *. Single * indicates significant difference compared to all other groups at the same time point.
-
Figure 5—source data 1
Quantification of regenerating myoblasts upon acute injury, qPCR results and number of myogenic committed cells in TA at day 0,2 and 4.
- https://cdn.elifesciences.org/articles/77610/elife-77610-fig5-data1-v2.xlsx
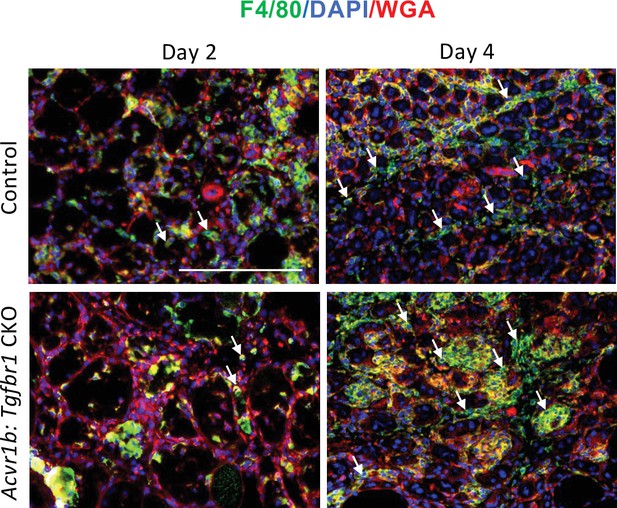
Number of macrophages in TA of Acvr1b:Tgfbr1 CKO animals was increased 4 days post injury.
Number of macrophages (F4/80+, green) in control and Acvr1b:Tgfbr1 CKO animals on days 2 and 4 post injury. Nuclei were stained by DAPI (blue) and ECM of muscle were stained by WGA (red). Scale bar = 100 μm. N = 5–7.
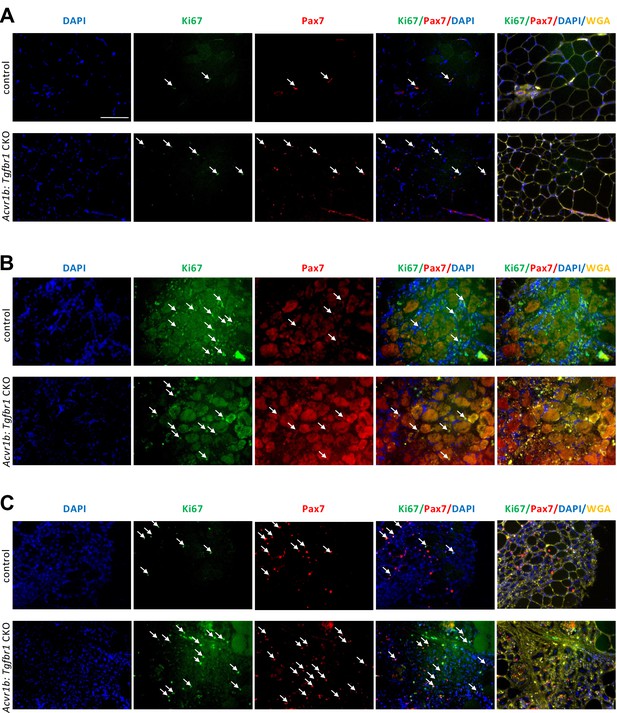
Increased number of proliferating cells and satellite cells in TA of Acvr1b:Tgfbr1 CKO animals in absence of injury and 4 days post injury.
(A) In absence of injury, proliferating cells (Ki67+, green, white arrows) and muscle satellite cells (Pax7+, red, white arrows) were shown by IF staining in low oxidative area of TA, where regeneration patches were found. Two (B) and 4 (C) days post injury, more Ki67+ and Pax7+ cells (white arrows) infiltrated in injured area. Nuclei were stained by DAPI (blue) and ECM of muscle were stained by WGA (yellow). Scale bar = 100 μm. N = 5–7.
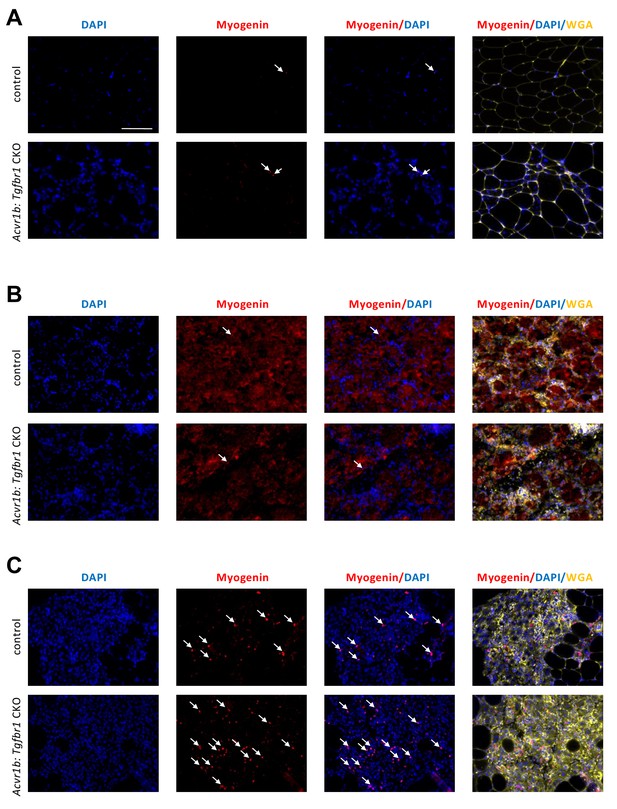
Increased number of differentiating muscle cells in TA of Acvr1b:Tgfbr1 CKO animals 4 days post injury.
On day 0 (A) and 2 days (B) post injury, the number of differentiating myoblasts (myogenin+, red, white arrows) was of no difference between groups. Four days (C) post injury, more Myogenin+ cells were found in injured area of Acvr1b:Tgfbr1 CKO animals. Nuclei were stained by DAPI (blue) and ECM of muscle were stained by WGA (yellow). Scale bar = 100 μm. N = 5–7.
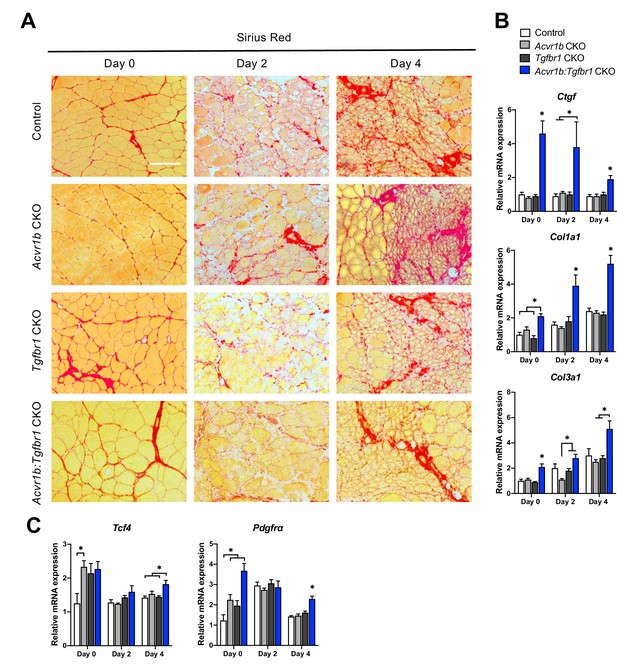
Relative mRNA expression levels of ECM components were enhanced in Acvr1b:Tgfbr1 CKO mice.
(A) Sirius Red staining shows collagen deposition in absence (day 0) or presence of CTX injection after 2, and 4 days (scale bar = 100 μm). (B, C) Relative gene expression in TA muscle in absence (day 0) or presence of CTX injection after 2 and 4 days. Results are presented as mean + SEM. N = 5–8 mice, *: p < 0.05. Significant differences between individual groups are indicated by lines with a *. Single * indicates significant difference compared to all other groups at the same time point.
-
Figure 6—source data 1
Quantification of qPCR for extracellular matrix genes in TA at day 0,2 and 4.
- https://cdn.elifesciences.org/articles/77610/elife-77610-fig6-data1-v2.xlsx
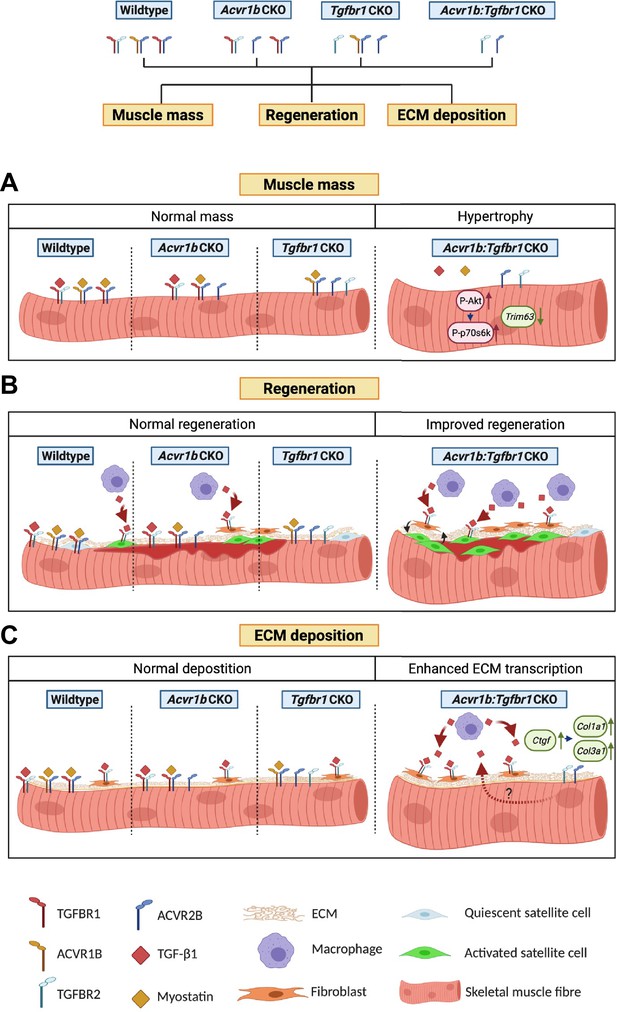
Schematic diagram of the effects of single or combined muscle-specific knockout of Tgfbr1 and/or Acvr1b receptors on muscle hypertrophy, regeneration, and expression of ECM components.
(A) Myofibre size is not affected after individual knockout of Acvr1b or Tgfbr1, which indicates that these receptors have redundant effects on muscle size and that myostatin signals via both receptors to control muscle mass. Simultaneous knockout of both Acvr1b and Tgfbr1 inhibits signaling of TGF-β, myostatin and activin A and stimulates protein synthesis via the Akt/mTOR/p70S6K pathway, while inhibiting protein breakdown through repression of Trim63 levels, resulting in substantial muscle hypertrophy. (B) Upon acute injury, simultaneous knockout of combined Acvr1b and Tgfbr1 accelerates early muscle regeneration, as observed by increased myogenic gene expression as well as increased CSA of regenerating myofibres. An increased number of SCs likely contributes to these effects. (C) Simultaneous myofibre-specific knockout of Acvr1b and Tgfbr1 induces mRNA expression of ECM components. These effects are likely caused by enhanced TGF- β1 signaling in fibroblasts. Schematic is created using BioRender.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (C57BL/6, males) | C57BL/6 | PMID:22564549 | # 025750 | Jackson Laboratory, Bar Harbor, ME, USA |
| Genetic reagent (Transfected construct (Mus musculus)) | HSA-Cre | PMID:22564549 | # 025750 | Jackson Laboratory, Bar Harbor, ME, USA |
| Genetic reagent (Transfected construct (Mus musculus)) | Acvr1bfl/fl | PMID:23109354 | Cancer Research Center of Lyon, French Institute of Health and Medical Research | |
| Genetic reagent (Transfected construct (Mus musculus)) | Tgfbr1fl/fl | PMID:11285230 | Leiden University Medical Center | |
| Antibody | Anti-phospho-Smad2 (Ser465/467) (138D4) (Rabbit monoclonal) | Cell Signaling Technology | Cat# 3108, RRID:AB_490941 | WB (1:500) |
| Antibody | Anti-phospho-Smad3 (Ser423/425) (C25A9) (Rabbit monoclonal) | Cell Signaling Technology | Cat# 9520, RRID:AB_10203253 | WB (1:500) |
| Antibody | Anti-Smad2/3 (Mouse monoclonal) | Cell Signaling Technology | Cat# 610843, RRID: AB_398162 | WB (1:500) |
| Antibody | Anti-phospho-AKT (Ser473) (Rabbit monoclonal) | Cell Signaling Technology | Cat# 9271, RRID:AB 329825 | WB (1:1000) |
| Antibody | Anti-AKT (pan) (C67E7) (Rabbit monoclonal) | Cell Signaling Technology | Cat# 4691, RRID:AB_915783 | WB (1:2000) |
| Antibody | Anti-phospho-p70S6 Kinase (Thr389) (108D2) (Rabbit monoclonal) | Cell Signaling Technology | Cat# 9234, RRID:AB_2269803 | WB (1:2000) |
| Antibody | Anti-p70S6 Kinase (49D7) (Rabbit monoclonal) | Cell Signaling Technology | Cat# 2708, RRID:AB_390722 | WB (1:2000) |
| Antibody | Anti-Pan-Actin (Rabbit polyclonal) | Cell Signaling Technology | Cat# 4968, RRID:AB_2313904 | WB (1:2000) |
| Antibody | Anti-Rabbit Immunoglobulins/HRP (Goat polyclonal) | Dako, Agilent | Cat# P0448, RRID:AB_2617138 | WB (1:2000) |
| Antibody | Anti-Mouse IgG (H + L), HRP (Rabbit polyclonal) | Thermo Fisher Scientific | Cat# 31457, RRID:AB_228439 | WB (1:2000) |
| Antibody | Anti-MHC-I (Mouse monoclonal) | DSHB | Cat# BA-D5, RRID:AB_2235587 | IF (1 µg/mL) |
| Antibody | Anti-MHC-IIA (Mouse monoclonal) | DSHB | Cat# SC-71, RRID:AB_2147165 | IF (10 µg/mL) |
| Antibody | Anti-MHC-IIB (Mouse monoclonal) | DSHB | Cat# BF-F3, RRID:AB_2266724 | IF (1 µg/mL) |
| Antibody | Anti-MHC-IIX (Mouse monoclonal) | DSHB | Cat# 6H1, RRID:AB_1157897 | IF (1 µg/mL) |
| Antibody | Anti-embryonic myosin heavy chain (eMyHc) (Mouse monoclonal) | DSHB | Cat# F1.652, RRID:AB_528358 | IF (20 µg/mL) |
| Antibody | Anti-Pax7 (Mouse monoclonal) | DSHB | Cat# PAX7, RRID:AB_2299243 | IF (4 µg/mL) |
| Antibody | Anti-F4/80 (D4C8V) XP (Rabbit monoclonal) | Cell Signaling Technology | Cat# 30325, RRID:AB_2798990 | IF (0.5 µg/mL) |
| Antibody | Anti-Myogenin (Mouse monoclonal) | DSHB | Cat# f5d, RRID:AB_2146602 | IF (0.6 µg/mL) |
| Antibody | Anti-Ki67 (Rabbit monoclonal) | Cell Signaling Technology | Cat# 11882, RRID:AB_2687824 | IF (1:200) |
| Antibody | Anti-Mouse Alexa Fluor 647 IgG2b (Goat polyclonal) | Thermo Scientific | Cat# A-21242, RRID:AB_2535811 | IF (5 µg/mL) |
| Antibody | Anti-Mouse Alexa Fluor 488 IgG1 (Goat polyclonal) | Thermo Scientific | Cat# A-21121, RRID:AB_2535764 | IF (5 µg/mL) |
| Antibody | Anti-Mouse Alexa Fluor 647 IgM (Goat polyclonal) | Thermo Scientific | Cat# A21238, RRID:AB_1500930 | IF (5 µg/mL) |
| Antibody | Anti-Mouse Alexa Fluor 488 IgM (Goat polyclonal) | Thermo Scientific | Cat# A-21042, RRID:AB_141357 | IF (5 µg/mL) |
| Antibody | Anti-Mouse Alexa Fluor 488 IgG (H + L) (Goat polyclonal) | Thermo Scientific | Cat# A-11029, RRID:AB_2534088 | IF (4–5 µg/mL) |
| Antibody | Anti-rabbit IgG (H + L), F(ab')2 Fragment Alexa Fluor 488 Conjugate (Goat polyclonal) | Cell Signaling Technology | Cat# 4412, RRID:AB_1904025 | IF (5 µg/mL) |
| Sequence-based reagent | Rps13-F | This paper | PCR primers | CACGTGGCTGAAGTTGACG |
| Sequence-based reagent | Rps13-R | This paper | PCR primers | CAGGATTACACCTATCTGGGAGG |
| Sequence-based reagent | Rpl27-F | This paper | PCR primers | AGCCGTCATCGTGAAGAAC |
| Sequence-based reagent | Rpl27-R | This paper | PCR primers | GGGGATAGCGGTCAATTCC |
| Sequence-based reagent | Tgfbr1-F | This paper | PCR primers | CCTCGAGACAGGCCATTTGT |
| Sequence-based reagent | Tgfbr1-F | This paper | PCR primers | AGACGAAGCAGACTGGACCA |
| Sequence-based reagent | Acvr1b-F | This paper | PCR primers | TGCTGCGCCATGAAAACATC |
| Sequence-based reagent | Acvr1b-F | This paper | PCR primers | TGCCCACAATCTCCATATGCA |
| Sequence-based reagent | Tgfb1-F | This paper | PCR primers | GCTGACCCCCACTGATACG |
| Sequence-based reagent | Tgfb1-R | This paper | PCR primers | CCTGTATTCCGTCTCCTTGGTT |
| Sequence-based reagent | Mstn-F | This paper | PCR primers | GAGAATGGCCATGATCTTGCTG |
| Sequence-based reagent | Mstn-R | This paper | PCR primers | CTTCTAAAAAGGGATTCAGCCCATC |
| Sequence-based reagent | Igf1ea-F | This paper | PCR primers | GTGTTGCTTCCGGAGCTGTG |
| Sequence-based reagent | Igf1ea-R | This paper | PCR primers | CAATGTACTTCCTTCTGAGTC |
| Sequence-based reagent | Hgf-F | This paper | PCR primers | GATTATTGCCCTATTTCCCGTTGTG |
| Sequence-based reagent | Hgf-R | This paper | PCR primers | TGGCACAGGATATTACAGGATGG |
| Sequence-based reagent | Igf1ec-F | This paper | PCR primers | GGAGAAGGAAAGGAAGTACATTTG |
| Sequence-based reagent | Igf1ec-R | This paper | PCR primers | CCTGCTCCGTGGGAGGCT |
| Sequence-based reagent | Vegfa-F | This paper | PCR primers | CTGTAACGATGAAGCCCTGGAGTG |
| Sequence-based reagent | Vegfa-R | This paper | PCR primers | GGTGAGGTTTGATCCGCATGATCT |
| Sequence-based reagent | Pax7-F | This paper | PCR primers | TCCATCAAGCCAGGAGACA |
| Sequence-based reagent | Pax7-R | This paper | PCR primers | AGGAAGAAGTCCCACACAG |
| Sequence-based reagent | Myod-F | This paper | PCR primers | CATCCAGCCCGCTCCAAC |
| Sequence-based reagent | Myod-R | This paper | PCR primers | GGGCCGCTGTAATCCATCATGCC |
| Sequence-based reagent | Myog-F | This paper | PCR primers | CCCAACCCAGGAGATCATTT |
| Sequence-based reagent | Myog-R | This paper | PCR primers | GTCTGGGAAGGCAACAGACA |
| Sequence-based reagent | Myh3-F | This paper | PCR primers | CGCAGAATCGCAAGTCAATA |
| Sequence-based reagent | Myh3-R | This paper | PCR primers | CAGGAGGTCTTGCTCACTCC |
| Sequence-based reagent | Id1-F | This paper | PCR primers | ACCCTGAACGGCGAGATCA |
| Sequence-based reagent | Id1-R | This paper | PCR primers | TCGTCGGCTGGAACACAT |
| Sequence-based reagent | Fgf2-F | This paper | PCR primers | AAGCGGCTCTACTGCAAGAA |
| Sequence-based reagent | Fgf2-R | This paper | PCR primers | GTAACACACTTAGAAGCCAGCAG |
| Sequence-based reagent | Ccn2-F | This paper | PCR primers | CCACCCGAGTTACCAATGAC |
| Sequence-based reagent | Ccn2-R | This paper | PCR primers | GCTTGGCGATTTTAGGTGTC |
| Sequence-based reagent | Col1a1-F | This paper | PCR primers | ATGTTCAGCTTTGTGGACCT |
| Sequence-based reagent | Col1a1-R | This paper | PCR primers | CAGCTGACTTCAGGGATGT |
| Sequence-based reagent | Col3a1-F | This paper | PCR primers | AAGGACATCGAGGATTCCCTG |
| Sequence-based reagent | Col3a1-R | This paper | PCR primers | AGCCCTCAGATCCTCTTTCAC |
| Sequence-based reagent | Cd68-F | This paper | PCR primers | TCCCAACAAAACCAAGGTCCA |
| Sequence-based reagent | Cd68-R | This paper | PCR primers | GGCTCTGATGTAGGTCCTGTTT |
| Sequence-based reagent | Cd163-F | This paper | PCR primers | CGGCCCCATGAAGAGGTATC |
| Sequence-based reagent | Cd163-R | This paper | PCR primers | GACGGTTGACCCAGTTGTTG |
| Sequence-based reagent | Il1b-F | This paper | PCR primers | GCCACCTTTTGACAGTGATG |
| Sequence-based reagent | Il1b-R | This paper | PCR primers | CTTCTCCACAGCCACAATGA |
| Sequence-based reagent | Il6-F | This paper | PCR primers | GGAAATGAGAAAAGAGTTGTGC |
| Sequence-based reagent | Il6-R | This paper | PCR primers | GTACTCCAGAAGACCAGAGGA |
| Sequence-based reagent | Fbxo32-F | This paper | PCR primers | AGACTGGACTTCTCGACTGC |
| Sequence-based reagent | Fbxo32-R | This paper | PCR primers | TCAGCTCCAACAACAGCCTTACT |
| Sequence-based reagent | Trim63-F | This paper | PCR primers | CGTCCAGAGCGTGTGTCTCACTC |
| Sequence-based reagent | Trim63-R | This paper | PCR primers | GGGCTACCTTCCTCTCAAGTGC |
| Sequence-based reagent | Tcf4-F | This paper | PCR primers | GGAAAGCCCTAGCTTCGATCT |
| Sequence-based reagent | Tcf4-R | This paper | PCR primers | GGAGCCCACAGGAGTTGAA |
| Sequence-based reagent | Pdgfra-F | This paper | PCR primers | ACTTTTCACTCCGGGTATCGG |
| Sequence-based reagent | Pdgfra-R | This paper | PCR primers | CCCATAGCTCCTGAGACCTTC |
| Commercial assay or kit | RiboPure RNA Purification Kit | Thermo Fisher Scientific | AM1924 | |
| Commercial assay or kit | SuperScript VILO Mastermix | Thermo Fisher Scientific | 12023679 |





