A network of cytosolic (co)chaperones promotes the biogenesis of mitochondrial signal-anchored outer membrane proteins
Figures
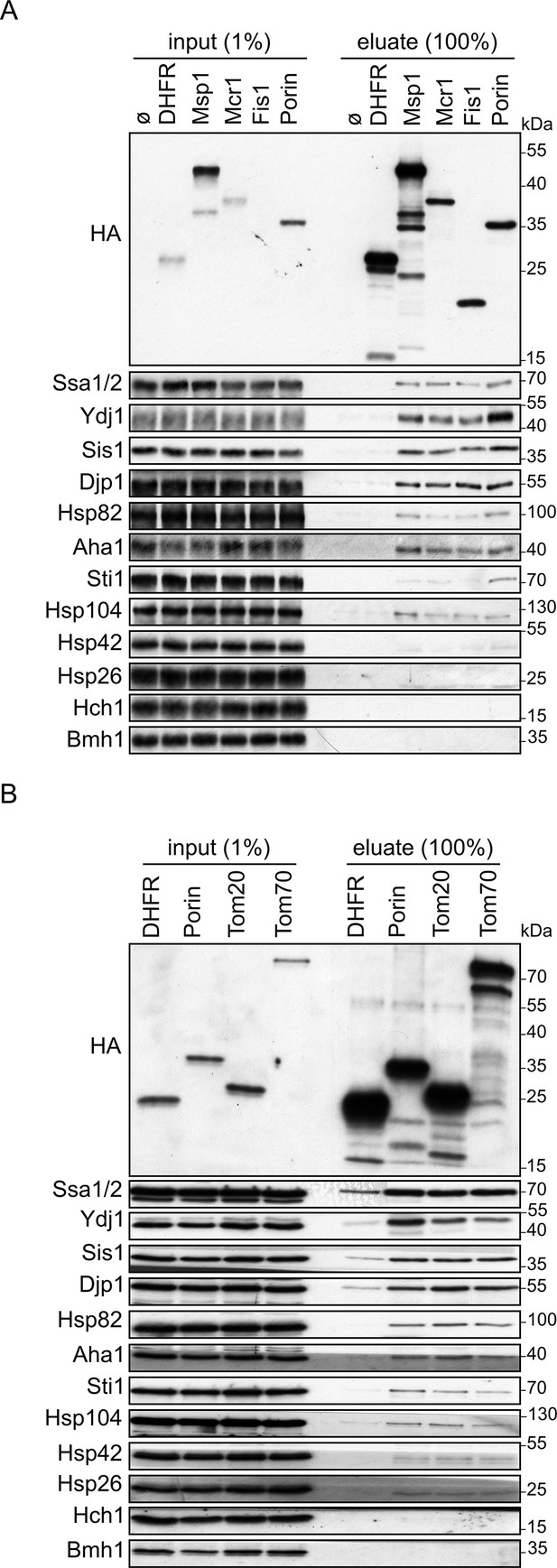
Cytosolic chaperones interact with newly synthesized signal-anchored proteins.
(A and B) In vitro translation reactions included yeast extracts without mRNA (Ø) or programmed with mRNA encoding HA-tagged variants of signal-anchored proteins (Msp1, Mcr1, Tom20, and Tom70), the tail-anchored protein Fis1, the β-barrel protein Porin, or, as a control, dihydrofolate reductase (DHFR). The reactions were subjected to a pull-down with anti-HA beads. Samples from the input (1%) and the eluates (100%) were analyzed by SDS-PAGE and immunodecoration with the indicated antibodies.
-
Figure 1—source data 1
Source data for Figure 1A.
- https://cdn.elifesciences.org/articles/77706/elife-77706-fig1-data1-v2.pdf
-
Figure 1—source data 2
Source data for Figure 1B.
- https://cdn.elifesciences.org/articles/77706/elife-77706-fig1-data2-v2.pdf
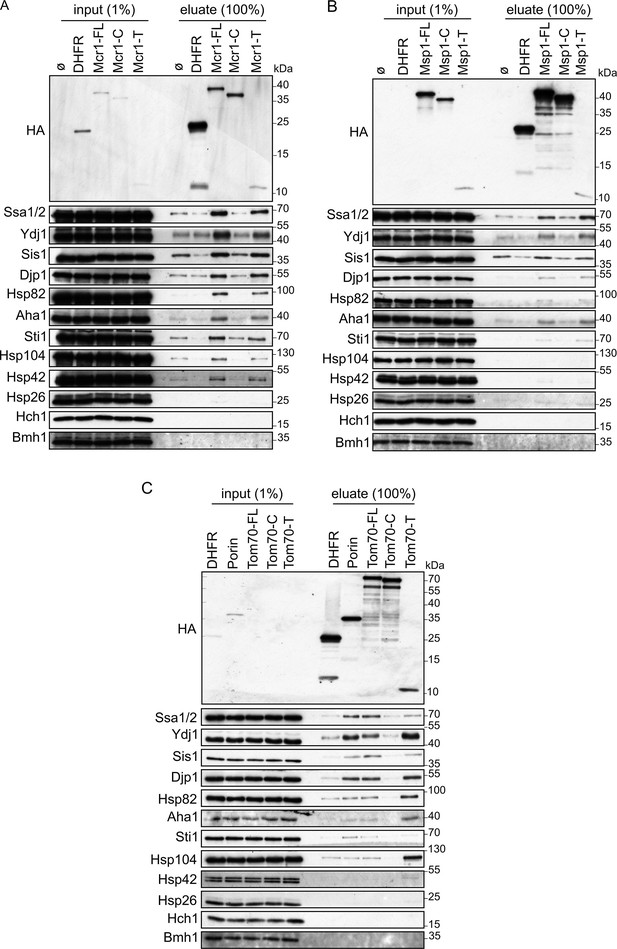
Cytosolic chaperones interact with newly synthesized signal-anchored proteins through their transmembrane segment.
(A–C) In-vitro translation reactions included yeast extracts without mRNA (Ø) or programmed with mRNA encoding HA-tagged versions of DHFR or the full length (FL), cytosolic domain (C) or transmembrane segment (T) of the SA proteins: Mcr1 (A), Msp1 (B) and Tom70 (C). The reactions were subjected to a pull-down with anti-HA beads. Samples from the input (1%) and the eluates (100%) were analyzed by SDS-PAGE and immunodecoration with the indicated antibodies.
-
Figure 2—source data 1
Source data for Figure 2A.
- https://cdn.elifesciences.org/articles/77706/elife-77706-fig2-data1-v2.pdf
-
Figure 2—source data 2
Source data for Figure 2B.
- https://cdn.elifesciences.org/articles/77706/elife-77706-fig2-data2-v2.pdf
-
Figure 2—source data 3
Source data for Figure 2C.
- https://cdn.elifesciences.org/articles/77706/elife-77706-fig2-data3-v2.pdf
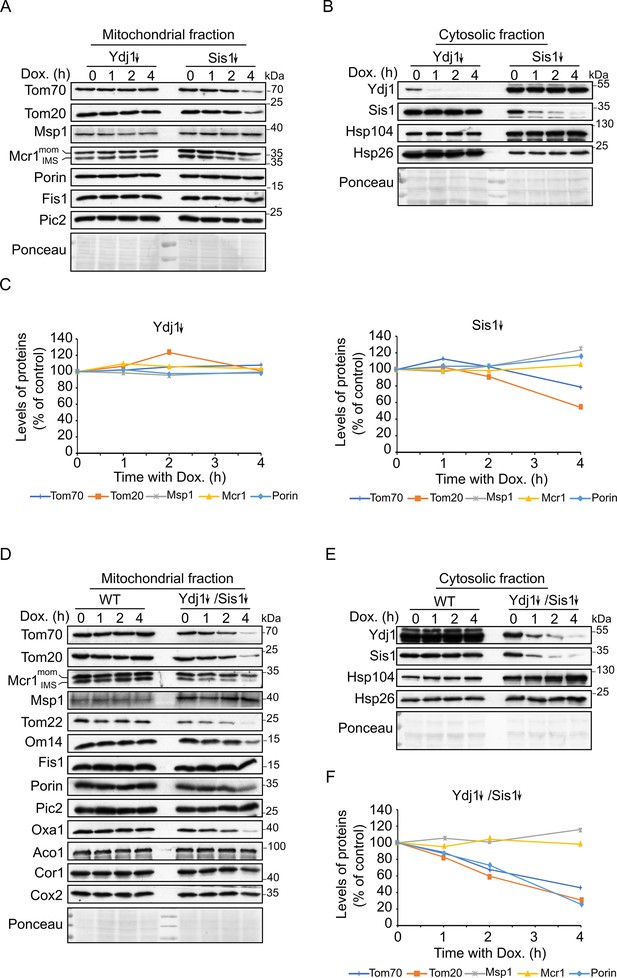
Depletion of both co-chaperones Ydj1 and Sis1 results in decreased steady-state levels of Tom20 and Tom70.
(A, B, D and E) Mitochondrial (A and D) and cytosolic (B and E) fractions were isolated from WT cells, cells depleted for either Ydj1 (Ydj1↓) or Sis1 (Sis1↓), or from cells double depleted for both co-chaperones (Ydj1↓Sis1↓). Cells were grown without doxycycline (time = 0) or in the presence of Dox for 1, 2, or 4 hr. Samples were analyzed by SDS-PAGE followed by immunodecoration with the indicated antibodies. (C and F) Intensities of the bands corresponding to the depicted proteins in the mitochondrial fractions from three independent experiments were quantified and normalized to Ponceau levels. The levels of the proteins in each depletion strain in the absence of doxycycline (time = 0) was set to 100%. Error bars represent ± SD.
-
Figure 3—source data 1
Source data for Figure 3A and B.
- https://cdn.elifesciences.org/articles/77706/elife-77706-fig3-data1-v2.pdf
-
Figure 3—source data 2
Source data for Figure 3D.
- https://cdn.elifesciences.org/articles/77706/elife-77706-fig3-data2-v2.pdf
-
Figure 3—source data 3
Source data for Figure 3E.
- https://cdn.elifesciences.org/articles/77706/elife-77706-fig3-data3-v2.pdf
-
Figure 3—source data 4
Raw data for plot in Figure 3C.
- https://cdn.elifesciences.org/articles/77706/elife-77706-fig3-data4-v2.xlsx
-
Figure 3—source data 5
Raw data for plot in Figure 3F.
- https://cdn.elifesciences.org/articles/77706/elife-77706-fig3-data5-v2.xlsx
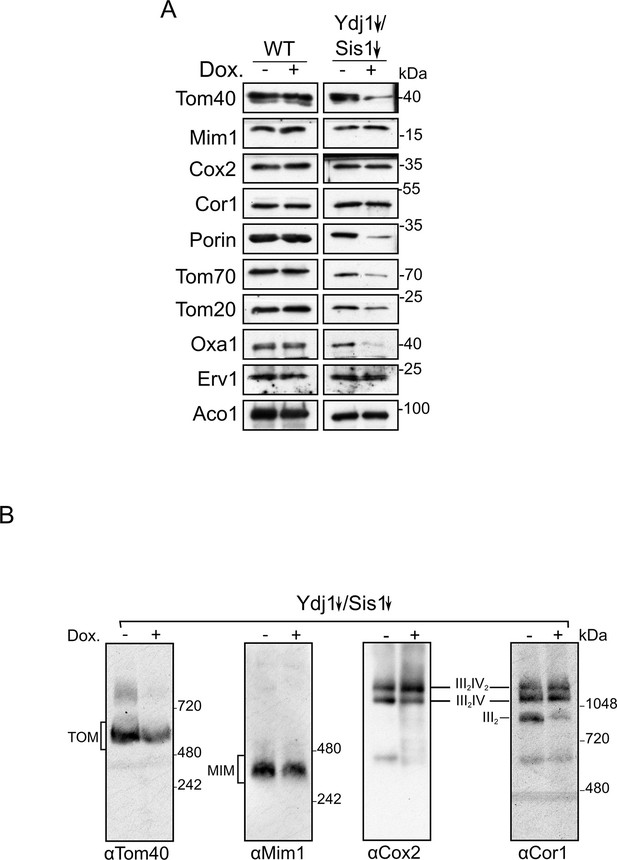
Depletion of both Ydj1 and Sis1 does not alter the assembly of the respiratory chain complexes or of the import machineries.
(A) Mitochondrial fractions were isolated from either WT or Ydj1 and Sis1 double depleted (Ydj1↓Sis1↓) cells, after being grown for 4 hr in the absence (-) or in the presence (+) of Dox. Samples were analyzed by SDS-PAGE followed by immunodecoration with the indicated antibodies. (B) Mitochondria isolated as in (A) were lysed in 1% Digitonin and subjected to 4–12% BN-BAGE. Proteins were analyzed by immunodecoration with antibodies against Tom40, Mim1, Cox2 of complex IV, and Cor1 of complex III. The migration of various supracomplexes is indicated.
-
Figure 3—figure supplement 1—source data 1
Source data for Figure 3—figure supplement 1A.
- https://cdn.elifesciences.org/articles/77706/elife-77706-fig3-figsupp1-data1-v2.pdf
-
Figure 3—figure supplement 1—source data 2
Source data for Figure 3—figure supplement 1B.
- https://cdn.elifesciences.org/articles/77706/elife-77706-fig3-figsupp1-data2-v2.pdf
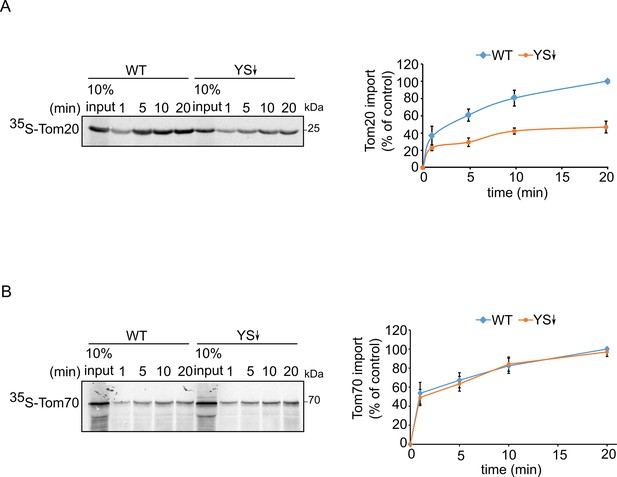
Signal-anchored proteins show variable dependence on Ydj1 and Sis1.
Radiolabeled Tom20 (A) or Tom70 (B) were translated in yeast extract from either WT or Ydj1 and Sis1 depleted cells (YS↓). The radiolabeled proteins were incubated with WT mitochondria for the indicated time points (1, 5, 10, and 20 min). After import, mitochondria were subjected to alkaline extraction and the pellet was analyzed by SDS-PAGE and autoradiography. Right panels: Intensities of the bands corresponding to Tom20 and Tom70 were quantified. The intensities of the bands corresponding to import from WT yeast extract after 20 min were set to 100%. The graph represents the mean values ± SD of three independent experiments.
-
Figure 4—source data 1
Source data for Figure 4A and B.
- https://cdn.elifesciences.org/articles/77706/elife-77706-fig4-data1-v2.pdf
-
Figure 4—source data 2
Raw data for plot in Figure 4A.
- https://cdn.elifesciences.org/articles/77706/elife-77706-fig4-data2-v2.xlsx
-
Figure 4—source data 3
Raw data for plot in Figure 4B.
- https://cdn.elifesciences.org/articles/77706/elife-77706-fig4-data3-v2.xlsx
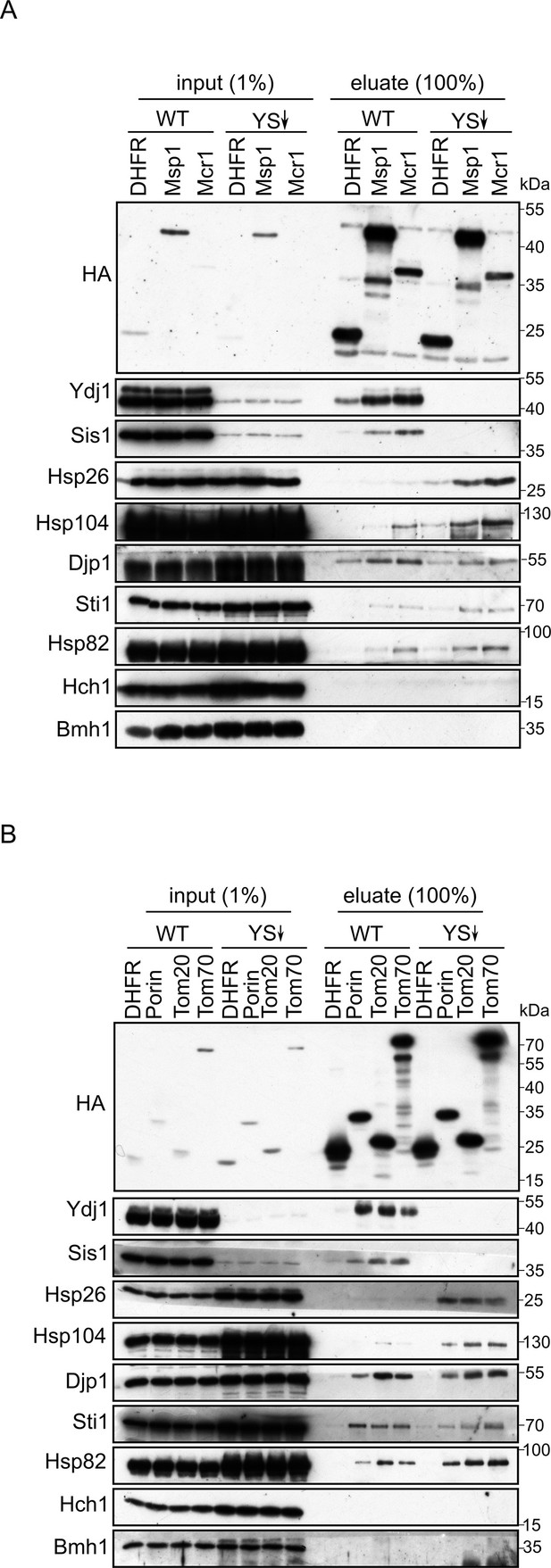
Depletion of Ydj1 and Sis1 can increase the risk for aggregation of newly synthesized proteins.
In-vitro translation reactions using yeast extracts from either WT cells or from cells depleted for both Ydj1 and Sis1 (YS↓) were programmed with mRNA encoding HA-tagged versions of the indicated proteins (A), DHFR, Msp1, and Mcr1; (B), DHFR, (Porin, Tom20, and Tom70). The reactions were subjected to a pull-down with anti-HA beads. Samples from the input (1%) and the eluates (100%) were analyzed by SDS-PAGE and immunodecoration with the indicated antibodies.
-
Figure 5—source data 1
Source data for Figure 5A.
- https://cdn.elifesciences.org/articles/77706/elife-77706-fig5-data1-v2.pdf
-
Figure 5—source data 2
Source data for Figure 5B.
- https://cdn.elifesciences.org/articles/77706/elife-77706-fig5-data2-v2.pdf
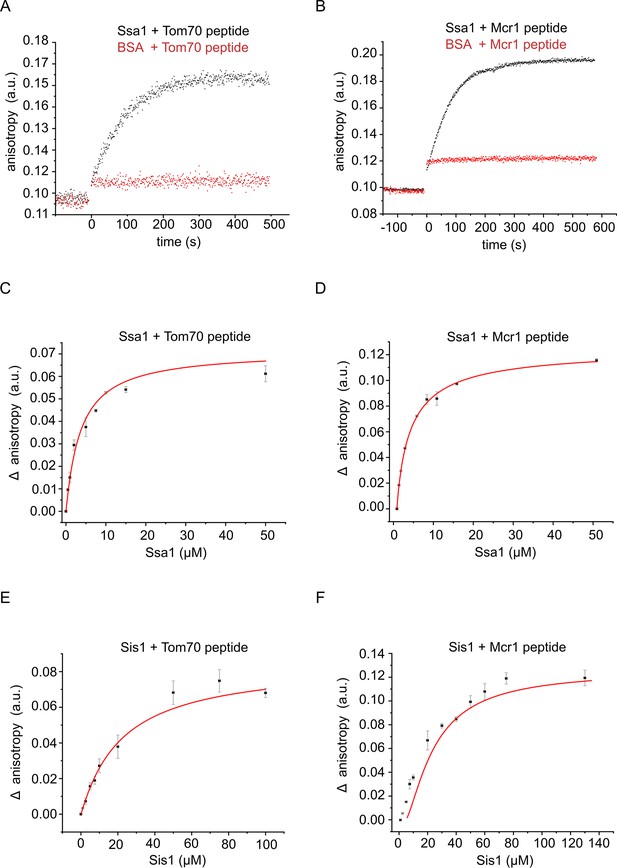
The hydrophobic segment of the signal-anchored proteins interacts with the Hsp70 chaperone and its co-chaperone Sis1.
(A and B) The fluorescence anisotropy of TAMRA-labeled peptides corresponding to the TMSs of either Tom70 (A) or Mcr1 (B) was measured in the presence of 10 µM of the Hsp70 Ssa1 (black circles) or 30 µM BSA, as a control (red circles). (C–F) For affinity determinations, the TMS-labelled peptides of either Tom70 (C and E) or Mcr1 (D and F) were mixed with the indicated concentrations of either Ssa1 (C and D) or Sis1 (E and F), and the difference in anisotropy (Δ anisotropy) between the bound and free peptide was plotted against the (co)chaperone concentrations.
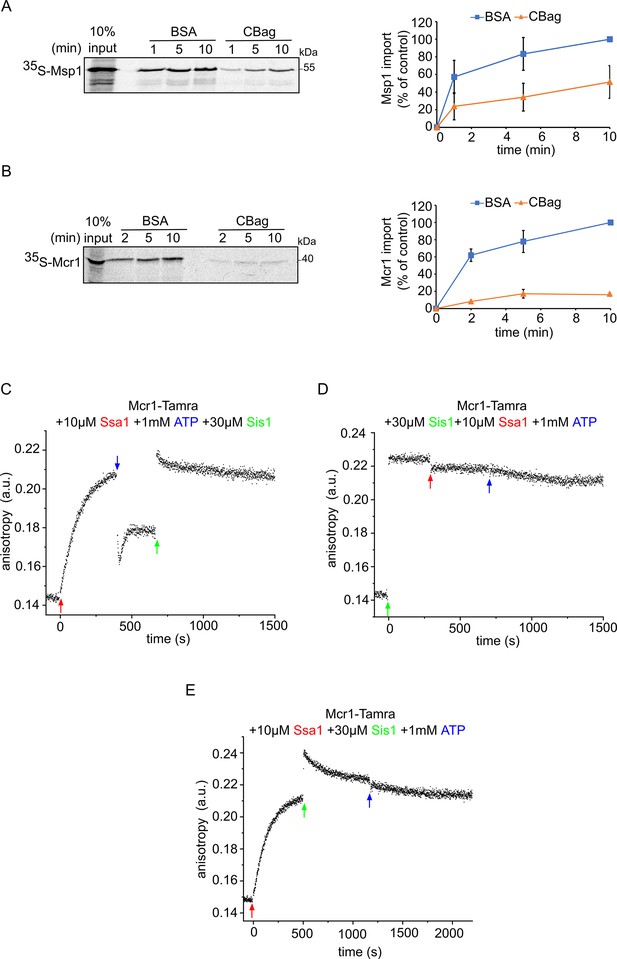
The Hsp70 chaperone Ssa1 is required for proper membrane integration of signal-anchored proteins.
(A and B) Left panels: Radiolabeled Msp1 (A) and Mcr1 (B) were translated in yeast extract from WT cells and subjected to in-vitro import assay using isolated mitochondria. Prior to the import, the yeast extract translation reaction was incubated with either CBag (Hsp70 inhibitor) or with BSA, as a control. After import for the indicated time periods, the samples were subjected to carbonate extraction and the pellets fraction were analysed by SDS-PAGE followed by autoradiography. Right panels: The bands corresponding to Msp1 and Mcr1 were quantified and the results of three independent experiments are presented as mean values ± SD. The intensities of the bands corresponding to import for 10 min in the presence of BSA were set to 100%. (C–E) The fluorescence anisotropy of TAMRA-labelled Mcr1-TMS peptide was measured while supplementing 10 µM Ssa1, 30 µM Sis1, and 1 mM ATP in the order indicated in the various panels.
-
Figure 7—source data 1
Source data for Figure 7A and B.
- https://cdn.elifesciences.org/articles/77706/elife-77706-fig7-data1-v2.pdf
-
Figure 7—source data 2
Raw data for plot in Figure 7A.
- https://cdn.elifesciences.org/articles/77706/elife-77706-fig7-data2-v2.xlsx
-
Figure 7—source data 3
Raw data for plot in Figure 7B.
- https://cdn.elifesciences.org/articles/77706/elife-77706-fig7-data3-v2.xlsx
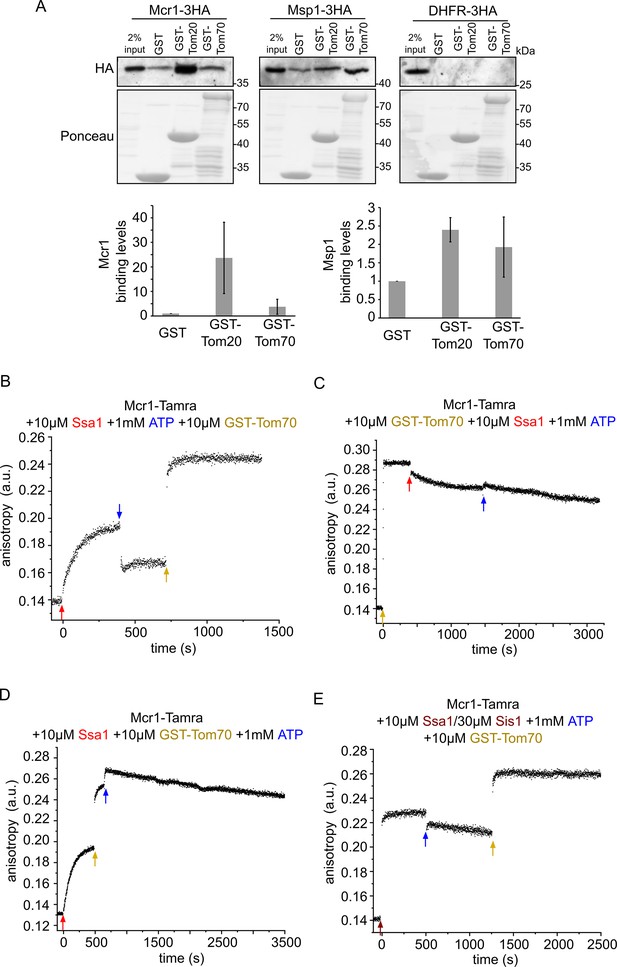
Newly synthesized signal-anchored proteins can be recognized by the cytosolic domains of the TOM receptors.
(A) HA-tagged versions of the signal-anchored proteins Mcr1 and Msp1 or of the control protein DHFR were freshly translated in yeast extract. Next, the newly translated proteins were mixed with GST alone or GST fused to the cytosolic domain of either Tom20 (GST-Tom20) or Tom70 (GST-Tom70) bound to glutathione beads. Input (2%) and eluate (100%) samples were subjected to SDS-PAGE. GST fusion proteins were detected by Ponceau staining whereas the HA-tagged proteins via immunodecoration against the HA-tag. Lower panels: Bands corresponding to Msp1-3HA and Mcr1-3HA from three independent experiments were quantified and the level of binding to GST alone was set as 1. Error bars represent ± SD. (B–D) Fluorescence anisotropy of TAMRA-labeled Mcr1 peptide was monitored after supplementing the reaction with 10 µM Ssa1, 1 mM ATP, or 10 µM GST-Tom70 in the indicated order. (E) As in panels B-D while the first addition was of 10 µM Ssa1 together with 30 µM Sis1, followed by addition of 1 mM ATP and then finally 10 µM GST-Tom70.
-
Figure 8—source data 1
Source data for Figure 8A.
- https://cdn.elifesciences.org/articles/77706/elife-77706-fig8-data1-v2.pdf
-
Figure 8—source data 2
Raw data for plots in Figure 8A.
- https://cdn.elifesciences.org/articles/77706/elife-77706-fig8-data2-v2.xlsx
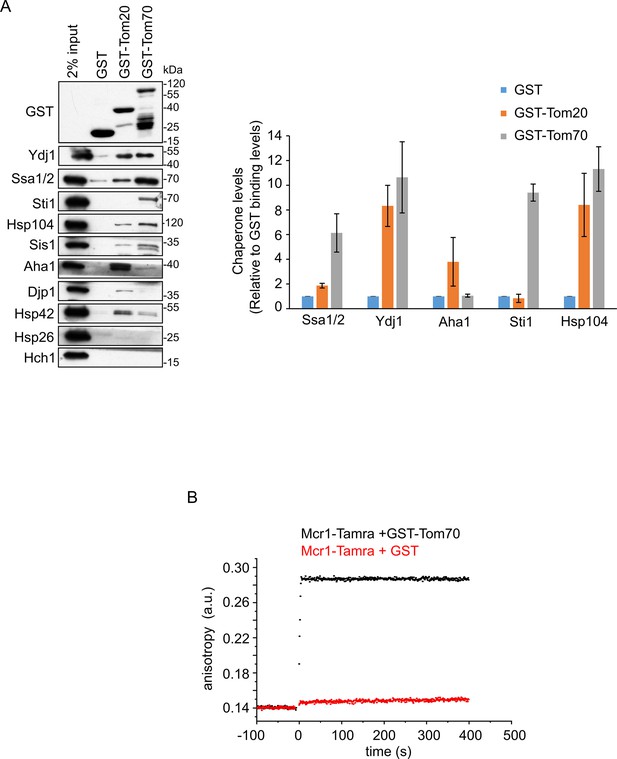
The TOM receptors interact with various chaperones.
(A) Left panel: Yeast extract was incubated with GST alone or with GST fused to the cytosolic domain of either Tom20 (GST-Tom20) or Tom70 (GST-Tom70). Samples of the input (2%) and the eluate (100%) were subjected to SDS-PAGE followed by immunodecoration with the indicated antibodies. Right panel: Bands representing the different (co)chaperones in the elution fractions were quantified and the results of three independent experiments are presented as mean values ± SD. The protein levels in the eluate using GST alone were set to 1. (B) Fluorescence anisotropy of TAMRA-labeled Mcr1 peptide was monitored after supplementing the reaction with either GST-Tom70 (black circles) or GST alone (red circles).
-
Figure 8—figure supplement 1—source data 1
Source data for Figure 8—figure supplement 1.
- https://cdn.elifesciences.org/articles/77706/elife-77706-fig8-figsupp1-data1-v2.pdf
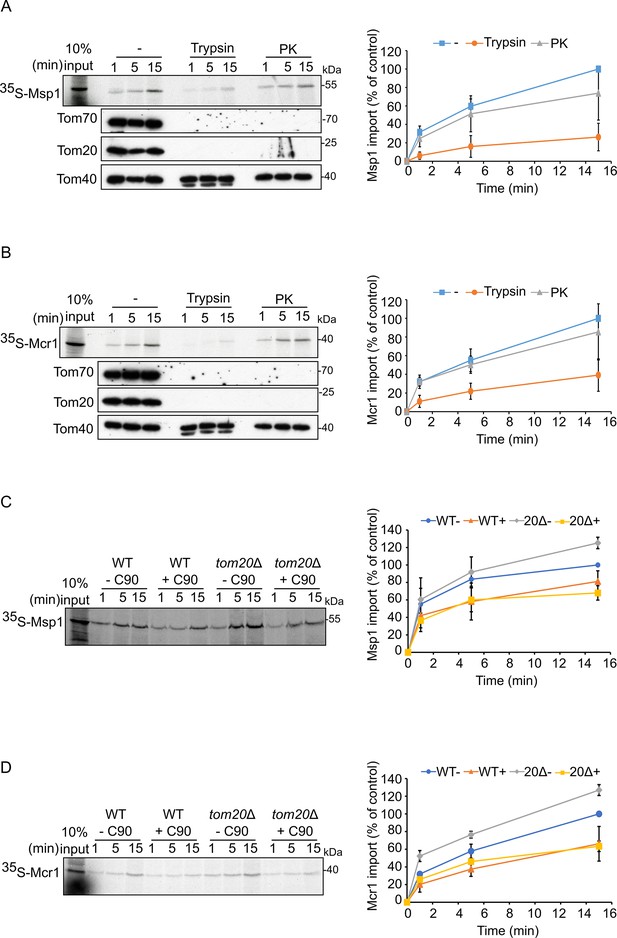
Tom70 and Tom20 may have offsetting function in mediating the biogenesis of Msp1 and Mcr1.
(A and B) Left panels: Radiolabeled Msp1 (A) and Mcr1 (B) were translated in yeast extract from WT cells and subjected to in vitro import assay using isolated mitochondria. Prior to the import reactions, isolated mitochondria were incubated for 30 min in the presence or absence of either trypsin or proteinase K (PK). After import for the indicated time periods, the samples were subjected to carbonate extraction and the pellet fractions were subjected to SDS-PAGE followed by autoradiography. To verify the activity of the proteases, the same membranes were immunodecorated with antibodies against the indicated proteins. Right panels: The bands corresponding to Msp1 or Mcr1 were quantified and the results of three independent experiments are presented as mean values ± SD. The intensities of the bands corresponding to import for 15 min in the absence of protease were set to 100%. (C and D) Left panels: Radiolabeled Msp1 (C) and Mcr1 (D) were translated in yeast extract from WT cells and subjected to in-vitro import assay using mitochondria isolated from either WT or tom20Δ cells. Prior to the import reactions, mitochondria were incubated in the presence or absence of 20 μM C90 (blocker of Tom70). After import for the indicated time points, the samples were subjected to carbonate extraction and the pellet fractions were analyzed by SDS-PAGE followed by autoradiography. Right panels: The bands corresponding to Msp1 or Mcr1 were quantified and the results of three independent experiments are presented as mean values ± SD. The intensities of the bands corresponding to import for 15 min in the absence of C90 were set to 100%.
-
Figure 9—source data 1
Source data for Figure 9A and B.
- https://cdn.elifesciences.org/articles/77706/elife-77706-fig9-data1-v2.pdf
-
Figure 9—source data 2
Source data for Figure 9C and D.
- https://cdn.elifesciences.org/articles/77706/elife-77706-fig9-data2-v2.pdf
-
Figure 9—source data 3
Raw data for plots in Figure 9A and B.
- https://cdn.elifesciences.org/articles/77706/elife-77706-fig9-data3-v2.xlsx
-
Figure 9—source data 4
Raw data for plots in Figure 9C and D.
- https://cdn.elifesciences.org/articles/77706/elife-77706-fig9-data4-v2.xlsx
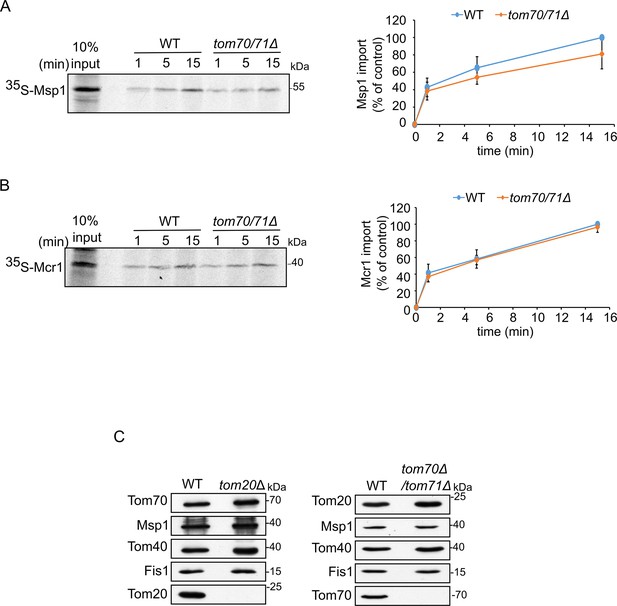
Biogenesis of SA proteins is not affected by the single deletion of a TOM receptor.
(A and B) Left panels: Radiolabeled Msp1 (A) or Mcr1 (B) were translated in yeast extract from WT cells and subjected to in vitro import assay using mitochondria isolated from either WT or tom70/71Δ strain. Right panels: The bands corresponding to Msp1 or Mcr1 were quantified and the results of three independent experiments are presented as mean values ± SD. The intensities of the bands corresponding to import for 15 min into control organelles were set to 100%. (C) Mitochondria isolated from either tom20Δ or tom70/71Δ deletion cells and their respective parental strain were analyzed by SDS-PAGE and immunodecoration with antibodies against the indicated proteins.
-
Figure 9—figure supplement 1—source data 1
Source data for Figure 9—figure supplement 1A and B.
- https://cdn.elifesciences.org/articles/77706/elife-77706-fig9-figsupp1-data1-v2.pdf
-
Figure 9—figure supplement 1—source data 2
Source data for Figure 9—figure supplement 1C.
- https://cdn.elifesciences.org/articles/77706/elife-77706-fig9-figsupp1-data2-v2.pdf
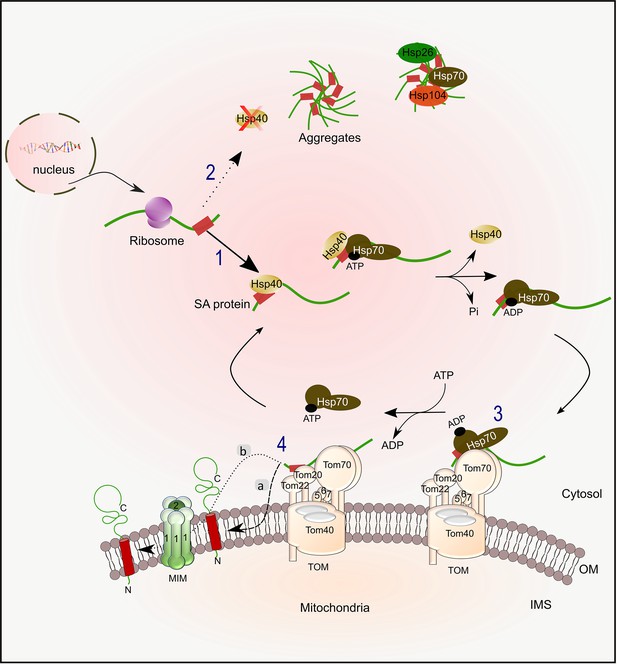
Working model for the biogenesis of SA proteins.
After SA proteins get synthesized on cytosolic ribosomes, they can associate with Hsp40 chaperones (like Ydj1 and Sis1)(1). Upon depletion of Hsp40 chaperones, newly synthesized SA proteins might tend to form aggregates, which can then associate with disaggregases chaperones such as Hsp104 and Hsp26 (2). Hsp40 chaperones drive the transfer of the newly synthesized protein to Hsp70 chaperone (Ssa1/2) by facilitating the conversion of Hsp70 from its ATP form to the ADP one that has a higher affinity for polypeptides. Next, the protein-chaperone complex is recognized by Tom70 receptor (3), followed by disassociation of the chaperone. Subsequently to the recognition by Tom70, which may involve also Tom20, the substrate is then inserted into the OM, either through an unassisted route (4 a), or via a pathway which is facilitated by the MIM complex (4b).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Saccharomyces cerevisiae) | WT | This paper | W303α | |
| Strain, strain background (Saccharomyces cerevisiae) | WT | This paper | JSY7452 | |
| Strain, strain background (Saccharomyces cerevisiae) | tetO7-Ubi-L-ydj1 | This paper | YMK120α, YDJ1::tetO7-Ubiquitin-Leu-YDJ1 KanMX4 | |
| Strain, strain background (Saccharomyces cerevisiae) | tetO7-Ubi-L-sis1 | This paper | YMK120α, SIS1::tetO7-Ubiquitin-Leu-SIS1 His3MX | |
| Strain, strain background (Saccharomyces cerevisiae) | tetO7-Ubi-L-ydj1/sis1 | This paper | YMK120α, tetO7-Ubi-Leu-SIS1:HisMX3a; tetO7-Ubi-Leu-YDJ1:KanMX4 | |
| Strain, strain background (Saccharomyces cerevisiae) | tom20Δ | This paper | W303α, TOM20::HIS3 | |
| Strain, strain background (Saccharomyces cerevisiae) | tom70/71Δ | This paper | JSY7452, TOM70::TRP1, TOM71::HIS3 | |
| Sequence-based reagent | Msp1 Fwd | This paper | PCR primers | GGGGGATCCATGTCTCGCAAA TTTGATTTAAAAACGATTACT GATCTTT |
| Sequence-based reagent | Msp1 Rev | This paper | PCR primers | GGGAAGCTTATCAAGAGGTTGA GATGACAACGTACTTG |
| Sequence-based reagent | yk Msp1 Fwd | This paper | PCR primers | GGGGGATCCAAAAAAATGT CTCGCAAATTTGATTTAAAA ACGATTACTGATCTTT |
| Sequence-based reagent | Yk Msp1 Rev | This paper | PCR primers | GGGAAGCTTTTAATCAAGA GGTTGAGATGACAAC |
| Sequence-based reagent | yk Msp1-3HA Fwd | This paper | PCR primers | CACACGAGCTCAAAAAAA TGTCTCGCAAATTTGATTTAA AAACG |
| Sequence-based reagent | yk Msp1-3HA Rev | This paper | PCR primers | CACACGGATCCCCATCAAG AGGTTGAGATGACAACGTAC |
| Sequence-based reagent | yk Mcr1-3HA Fwd | This paper | PCR primers | GGGGAATTCAAAAAAATGT TTTCCAGATTATCCAGATCTCACTCAAAAGC |
| Sequence-based reagent | yk Mcr1-3HA Rev | This paper | PCR primers | GGGCCCGGGAAATTTG AAAACTTGGTCCTTGGAGTAG CCC |
| Sequence-based reagent | yk Tom20-3HA Fwd | This paper | PCR primers | GGGGGTACCAAAAAAATG TCCCAGTCGAACCCTATCT TAC |
| Sequence-based reagent | yk Tom20-3HA Rev | This paper | PCR primers | GGGGGATCCGGGTCA TCGATATCGTTAGCTTCAGC |
| Sequence-based reagent | yk Tom70-3HA Fwd | This paper | PCR primers | GGGGGTACCAAAAAAAT GAAGAGCTTCATTACAAGGA ACAAGAC |
| Sequence-based reagent | yk Tom70-3HA Rev | This paper | PCR primers | GGGGGATCCGGCATTAA ACCCTGTTCGCGTAATTTAGC |
| Sequence-based reagent | yk Msp1-TMD-3HA Fwd | This paper | PCR primers | GGGGAATTCAAAAAAA TGTCTCGCAAATTTGATTTAA AAACG |
| Sequence-based reagent | yk Msp1-TMD-3HA Rev | This paper | PCR primers | GGGGGATCCCCGTT GAGTAGCCGACTGACCA |
| Sequence-based reagent | yk Msp1-CD-–3HA Fwd | This paper | PCR primers | CACACGAGCTCAAAAAAA TGGATGTTGAATCAGGACCGTTATCAGG |
| Sequence-based reagent | yk Msp1-CD-–3HA Rev | This paper | PCR primers | CACACGGATCCCCATCAAG AGGTTGAGATGACAACGTAC TTGTAGC |
| Sequence-based reagent | yk Mcr1-TMD-3HA Fwd | This paper | PCR primers | CACACGAATTCAAAAAAA TGTTTTCCAGATTATCCAG ATCTC |
| Sequence-based reagent | yk Mcr1-TMD-3HA Rev | This paper | PCR primers | CACACCCCGGGGACAAAGG AATGTTGGTTACG GTTT |
| Sequence-based reagent | yk Mcr1-CD-3HA Fwd | This paper | PCR primers | CACACGAATTCAAAAAAAT GCATTCCTTTGTCTTCAATG AATC |
| Sequence-based reagent | yk Mcr1-CD-3HA Rev | This paper | PCR primers | CACACCCCGGGAAATTTGA AAACTTGGTCCTTGGAGTAG |
| Sequence-based reagent | yk Tom20-TMD-3HA Fwd | This paper | PCR primers | GGGGGTACCAAAAAAATGT CCCAGTCGAACCCTATCTTAC |
| Sequence-based reagent | yk Tom20-TMD-3HA Rev | This paper | PCR primers | GGGGGATCCGGGTCAAA GTAGATAGCATAACCGGTG |
| Sequence-based reagent | yk Tom20-CD-3HA Fwd | This paper | PCR primers | GGGGGTACCAAAAAAAT GAGAAATAGCCCGCAATTC AGGAA |
| Sequence-based reagent | yk Tom20-CD-3HA Rev | This paper | PCR primers | GGGGGATCCGGGTCATC GATATCGTTAGCTTCAGC |
| Sequence-based reagent | yk Tom70-TMD-3HA Fwd | This paper | PCR primers | GGGGGTACCAAAAAAA TGAAGAGCTTCATTACAAGGA ACAAGAC |
| Sequence-based reagent | yk Tom70-TMD-3HA Rev | This paper | PCR primers | GGGGGATCCGGCAATTGGT TGTAATAATAGTAGGCACC |
| Sequence-based reagent | yk Tom70-CD-3HA Fwd | This paper | PCR primers | GGGGGTACCAAAAAAATGC AACAACAACAACGAGGAAAAAAGAACAC |
| Sequence-based reagent | yk Tom70-CD-3HA Rev | This paper | PCR primers | GGGGGATCCGGCATTAAACC CTGTTCGCGTAATTTAGC |
| Sequence-based reagent | tetO7-Ubi-L-Ydj1 Fwd | Jores et al., 2018 | PCR primers | CATATCTTTTGATAGAACATA ATTAAAAATTATCCAAACTGA ATTCTACACAGTATAGCGACC AGCATTCACATACG |
| Sequence-based reagent | tetO7-Ubi-L-Ydj1 Rev | Jores et al., 2018 | PCR primers | GTGGCAGTTACTGGAACACC TAGAATATCGTAAAACTTAG TTTCTTTAACCAAACCACCTC TCAATCTCAAGACCAAG |
| Sequence-based reagent | tetO7-Ubi-L-Sis1 Fwd | Jores et al., 2018 | PCR primers | GGATAAGTTGTTTGCATTTTA AGATTTTTTTTTTAATACATT CACATCAACAGTATAGCGAC CAGCATTCACATACG |
| Sequence-based reagent | tetO7-Ubi-L-Sis1 Rev | Jores et al., 2018 | PCR primers | TTAGCACTTGGAGATACT CCAAGTAAATCATAAAGTTT TGTCTCCTTGACCAAACCACC TCTCAATCTCAAGACCAAG |
| Recombinant DNA reagent | pGEM4polyA-3HA (plasmid) | Jores et al., 2018 | C-terminal 3 x HA-tag | |
| Recombinant DNA reagent | pGEM4polyA-yk-DHFR-3HA (plasmid) | Jores et al., 2018 | Yeast kozak sequence (AAAAAAATG) DHFR-3 ×HA-tag | |
| Recombinant DNA reagent | pGEM4polyA-yk-Porin-3HA (plasmid) | Jores et al., 2018 | Yeast kozak sequence (AAAAAAATG) Porin-3 ×HA-tag | |
| Recombinant DNA reagent | pGEM4polyA-yk-Fis1-3HA (plasmid) | This paper | Yeast kozak sequence (AAAAAAATG) Fis1−3×HA-tag | |
| Recombinant DNA reagent | pGEM4polyA-yk-Msp1-3HA (plasmid) | This paper | Yeast kozak sequence (AAAAAAATG) Msp1−3×HA-tag | |
| Recombinant DNA reagent | pGEM4polyA-yk-Mcr1-3HA (plasmid) | This paper | Yeast kozak sequence (AAAAAAATG) Mcr1−3×HA-tag | |
| Recombinant DNA reagent | pGEM4polyA-yk-Tom20-3HA (plasmid) | This paper | Yeast kozak sequence (AAAAAAATG) Tom20−3×HA-tag | |
| Recombinant DNA reagent | pGEM4polyA-yk-Tom70-3HA (plasmid) | This paper | Yeast kozak sequence (AAAAAAATG) Tom70−3×HA-tag | |
| Recombinant DNA reagent | pGEM4polyA-yk-Msp1(33-363)–3 HA (plasmid) | This paper | Yeast kozak sequence (AAAAAAATG) Msp1(1-363)–3×HA-tag | |
| Recombinant DNA reagent | pGEM4polyA-yk-Msp1(1-32)–3 HA (plasmid) | This paper | Yeast kozak sequence (AAAAAAATG) Msp1(1-32)–3×HA-tag | |
| Recombinant DNA reagent | pGEM4polyA-yk-Mcr1(35-302)–3 HA (plasmid) | This paper | Yeast kozak sequence (AAAAAAATG) Mcr1(35-302)–3×HA-tag | |
| Recombinant DNA reagent | pGEM4polyA-yk-Mcr1(1-39)–3 HA (plasmid) | This paper | Yeast kozak sequence (AAAAAAATG) Mcr1(1-39)–3×HA-tag | |
| Recombinant DNA reagent | pGEM4polyA-yk-Tom20(33-183)–3 HA (plasmid) | This paper | Yeast kozak sequence (AAAAAAATG) Tom20(33-183)–3×HA-tag | |
| Recombinant DNA reagent | pGEM4polyA-yk-Tom20(1-30)–3 HA (plasmid) | This paper | Yeast kozak sequence (AAAAAAATG) Tom20(1-30)–3×HA-tag | |
| Recombinant DNA reagent | pGEM4polyA-yk-Tom70(33-617)–3 HA (plasmid) | This paper | Yeast kozak sequence (AAAAAAATG) Tom70(33-617)–3×HA-tag | |
| Recombinant DNA reagent | pGEM4polyA-yk-Tom70(1-32)–3 HA (plasmid) | This paper | Yeast kozak sequence (AAAAAAATG) Tom70(1-32)–3×HA-tag | |
| Recombinant DNA reagent | pMK632His (plasmid) | Jores et al., 2018 | HIS3MX cassette tetO7-CYC1 promoter-Ubiquitin-Leucin-HA-tag | |
| Recombinant DNA reagent | pMK632Kan (plasmid) | Jores et al., 2018 | KanMX cassette tetO7-CYC1 promoter-Ubiquitin-Leucin-HA-tag | |
| Recombinant DNA reagent | pGEX4T1-GST (plasmid) | This paper | GST | |
| Recombinant DNA reagent | pGEX4T1-GST-Tom20(35-183) (plasmid) | This paper | Tom20(35-183) | |
| Recombinant DNA reagent | pGEX4T1-GST-Tom70 (46-617) (plasmid) | This paper | Tom70(46-617) | |
| Recombinant DNA reagent | pPROEX-HTa-cBag (plasmid) | Young et al., 2003 | His6-tag-TEV-human Bag-1M(151-263) | |
| Recombinant DNA reagent | pPROEX-HTa-(C90) (plasmid) | Young et al., 2003 | His6-tag-TEV-human Hsp90a(566-732) | |
| Antibody | Anti-Ssa1/2 (rabbit polyclonal) | Jores et al., 2018 | 1:20,000 | |
| Antibody | Anti-Ydj1 (rabbit polyclonal) | Jores et al., 2018 | 1:10,000 | |
| Antibody | Anti-Sis1 (rabbit polyclonal) | Jores et al., 2018 | 1:20,000 | |
| Antibody | Anti-Hsp26 (rabbit polyclonal) | Jores et al., 2018 | 1:4000 | |
| Antibody | Anti-Hsp104 (rabbit polyclonal) | Jores et al., 2018 | 1:25,000 | |
| Antibody | Anti-Hsp42 (rabbit polyclonal) | Jores et al., 2018 | 1:4000 | |
| Antibody | Anti-Hsp82 (rabbit polyclonal) | Jores et al., 2018 | 1:20,000 | |
| Antibody | Anti-Hch1 (rabbit polyclonal) | Jores et al., 2018 | 1:4000 | |
| Antibody | Anti-Bmh1 (rabbit polyclonal) | This paper | 1:1000 | |
| Antibody | Anti-Djp1 (rabbit polyclonal) | Lab of Ineke Braakman | 1:2000 | |
| Antibody | Anti-Sti1 (rabbit polyclonal) | This paper | 1:10,000 | |
| Antibody | Anti-Aha1 (rabbit polyclonal) | This paper | 1:2000 | |
| Antibody | Anti-Msp1 (rabbit polyclonal) | Lab of Toshiya Endo | 1:2000 | |
| Antibody | Anti-Mcr1 (rabbit polyclonal) | This paper | 1:2000 | |
| Antibody | Anti-Fis1 (rabbit polyclonal) | This paper | 1:1000 | |
| Antibody | Anti-Tom20 (rabbit polyclonal) | This paper | 1:4000 | |
| Antibody | Anti-Tom70 (rabbit polyclonal) | This paper | 1:5000 | |
| Antibody | Anti-Porin (rabbit polyclonal) | This paper | 1:6000 | |
| Antibody | Anti-Pic2 (rabbit polyclonal) | This paper | 1:2000 | |
| Antibody | Anti-HA (rat polyclonal) | Roche | #11867423001 | 1:1000 |
| Antibody | Anti-Cor1 (rabbit polyclonal) | This paper | 1:2000 | |
| Antibody | Anti-Mim1 (rabbit polyclonal) | This paper | 1:100 | |
| Antibody | Anti-Cox2 (rabbit polyclonal) | This paper | 1:2000 | |
| Antibody | Anti-Oxa1 (rabbit polyclonal) | This paper | 1:2000 | |
| Antibody | Anti-Erv1 (rabbit polyclonal) | Lab of Johannes Herrmann | 1:1000 | |
| Antibody | Anti-Aco1 (rabbit polyclonal) | This paper | 1:2000 | |
| Antibody | Anti-Tom22 (rabbit polyclonal) | This paper | 1:2000 | |
| Antibody | Anti-Om14 (rabbit polyclonal) | Lab of Thomas Becker | 1:2000 | |
| Antibody | Goat Anti-Rabbit IgG HRP conjugate | Bio-Rad | #1721019 | 1:10,000 |
| Antibody | Goat Anti-Rat IgG HRP conjugate | Abcam | #ab6845 | 1:2000 |
Additional files
-
Supplementary file 1
Proteins that co-purified with in vitro translated Msp1 or Mcr1.
(A) List of chaperones that were found in the elution fraction of either Msp1 or Mcr1 but not in the elution of mock pull-down (0). The iBAQ values of the indicated proteins are indicated. (B) A list of chaperones that were enriched in the elution fraction of either Msp1 or Mcr1 as compared to their levels in the elution from mock pull-down assay are indicated. The iBAQ value of each protein in the eluate of the mock pull-down was set to 1 and the relative values in the pull-down assays with wither Msp1 or Mcr1 are indicated.
- https://cdn.elifesciences.org/articles/77706/elife-77706-supp1-v2.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/77706/elife-77706-mdarchecklist1-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/77706/elife-77706-transrepform1-v2.docx





