Biphasic regulation of osteoblast development via the ERK MAPK–mTOR pathway
Figures
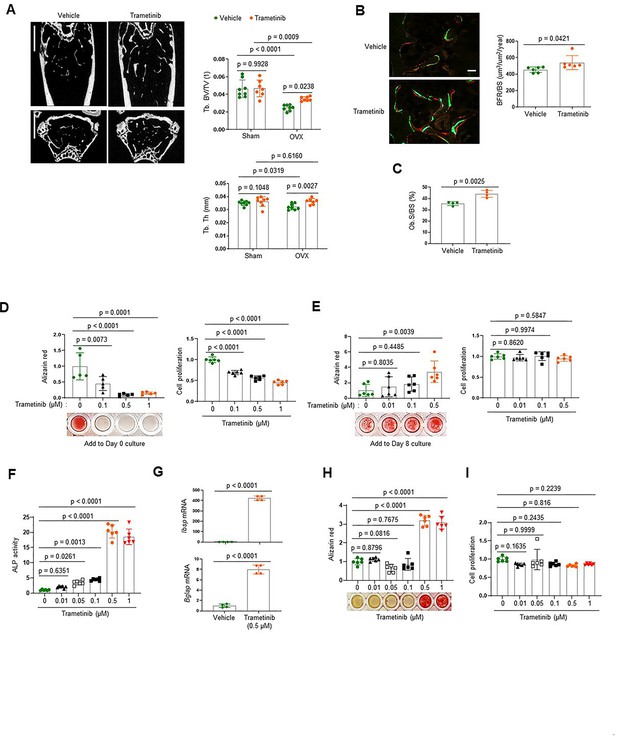
The extracellular signal-regulated kinase (ERK) mitogen-activated protein kinase (MAPK) pathway plays biphasic roles in osteogenesis.
(A–C) Twenty-week-old female mice were treated with vehicle or trametinib after either Sham or ovariectomy (OVX) surgery. Representative microCT images of the femoral bone (A, left) and quantification of relative trabecular bone volume/total volume (Tb. BV) and thickness (Tb. Th, A, right) are displayed. Alternatively, dynamic histomorphometry analysis of the femoral bones was performed to assess in vivo osteoblast activity in OVX group. Representative images of calcein/alizarin labeling (B, left) and quantification of bone formation rate (BFR)/bone surface (BS, B, right) and osteoblast surface (Ob.S)/bone surface BS, (C) are displayed. Scale bars, 1 mm (A), 50 µm (B). Human bone marrow-derived mesenchymal stromal cells (BMSCs) were treated with different doses of trametinib at days 0 (D) or 8 (E) of osteogenic culture. Mineralization and cell proliferation were assessed by alizarin red and alamar blue staining after 14 days of culture, respectively. (F–I) Mouse wildtype osteoblasts (Obs) were treated with different doses of trametinib at day 0 of osteogenic culture and 6 days later, alkaline phosphatase (ALP) activity (F) and osteogenic gene expression by real-time PCR (G) were assessed. Mineralization by alizarin red staining (H) and cell proliferation (I) by alamar blue staining were assessed after 18 days of culture. Data are representative of three independent experiments (A) [left], (B) [left], (D–I) or pooled from two experiments (A) [right], (B) [right], (C). A two-tailed unpaired Student’s t-test for comparing two groups (A–C, G) or ordinary one-way analysis of variance (ANOVA) with Dunnett’s multiple comparisons test (D–F, H, I) (A–I; error bars, standard deviation [SD] of biological replicates).
-
Figure 1—source data 1
Source data for Figure 1A–I.
- https://cdn.elifesciences.org/articles/78069/elife-78069-fig1-data1-v2.xlsx
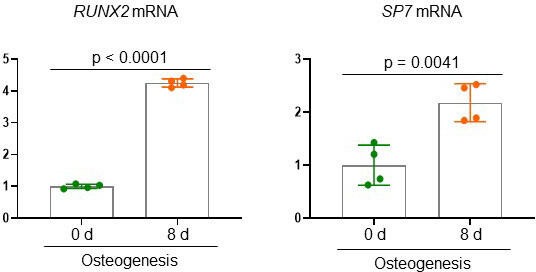
Osteogenic gene expression in human bone marrow-derived mesenchymal stromal cells (BMSCs).
Human BMSCs were cultured under osteogenic conditions, and mRNA levels of RUNX2 and SP7 were analyzed by RT-PCR at days 0 and 8 in culture. A two-tailed unpaired Student’s t-test for comparing two groups (error bars represent the standard deviation [SD] of biological replicates).
-
Figure 1—figure supplement 1—source data 1
Source data for Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/78069/elife-78069-fig1-figsupp1-data1-v2.xlsx
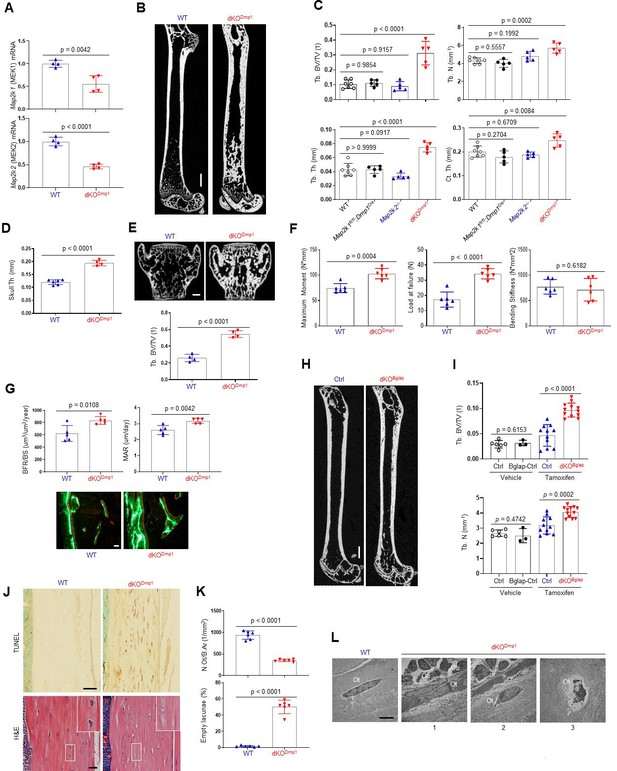
MEK1/2 deletion in mature osteoblasts promotes bone formation in mice.
(A) Map2k1 (MEK1) and Map2k2 (MEK2) mRNA levels in the tibial bones of Map2k1fl/fl;Map2k2+/+ (WT) and Map2k1fl/fl;Dmp1Cre;Map2k2−/− (dKODmp1) mice were measured by RT-PCR analysis. (B, C) MicroCT analysis showing femoral bone mass in 8-week-old Map2k1fl/fl;Map2k2+/+ (WT), Map2k1fl/fl;Dmp1Cre;Map2k2+/+ (Map2k1fl/fl;Dmp1Cre), Map2k1fl/fl;Map2k2−/− (Map2k2−/−), and Map2k1fl/fl;Dmp1Cre;Map2k2−/− (dKODmp1) mice. 3D reconstruction (B) and the relative quantification (C) are displayed. Trabecular bone volume/total volume (Tb. BV/TV), trabecular thickness (Tb. Th), trabecular number Tb. (N), and cortical thickness (Ct.Th). Scale bar, 1 mm (B). MicroCT analysis of 8-week-old WT and dKODmp1 skull (D) and vertebrae (E). Quantification of skull thickness Th (D), and trabecular bone mass (E, bottom) and 3D reconstruction of lumbar 4 (L4) vertebrae (E, top) are shown. Scale bar, 200 µm (E). (F) Biomechanical properties of 8-week-old WT and dKODmp1 femurs, including maximum moment, load at failure, and bending stiffness were quantified using three-point bending test. (G) Dynamic histomorphometry analysis of 8-week-old WT and dKODmp1 femurs. Quantification (top) and images of calcein/alizarin labeling (bottom) are displayed. BFR/BS, bone formation rate per bone surface; MAR, mineral apposition rate. Scale bar, 50 µm (bottom). (H, I) MicroCT analysis of femoral bone mass in 17-week-old Map2k1fl/fl;Map2k2−/− and Map2k1fl/fl;BglapCreErt;Map2k2−/− mice treated with vehicle or tamoxifen; vehicle-treated Map2k1fl/fl;Map2k2−/− (Ctrl) and Map2k1fl/fl;BglapCreErt;Map2k2−/− (Bglap-Ctrl) mice, tamoxifen-treated Map2k1fl/fl;Map2k2−/− (Ctrl) and Map2k1fl/fl;BglapCreErt;Map2k2−/− (dKOBglap) mice. 3D reconstruction (H) and the relative quantification (I) are displayed. Scale bar, 1 mm (H). (J) TUNEL (top) and hematoxylin and eosin (H&E) (bottom)-stained longitudinal sections of 8-week-old WT and dKODmp1 femurs. Scale bars, 50 µm (top) and 20 µm (bottom). (K) Numbers of osteocytes/bone area (N.Ot/B.Ar) and empty lacunae in the cortical bones of 8-week-old WT and dKODmp1 femurs. (L) Transmission electron microscopy (TEM) images of osteocytes in the cortical bones of 8-week-old WT and dKODmp1 femurs. 1, Ot in osteoids; 2, Ot in mineralized bone matrix; 3, apoptotic Ot in bone matrix. Ot, osteocyte; Oc, osteoclast; Ob, osteoblast. Scale bar, 5 µm. Data are representative of three independent experiments (A, B, E) [top], (G) [bottom], (H, J, L) or pooled from two experiments C, D, E [bottom], F, G [top], (I, K). A two-tailed unpaired Student’s t-test for comparing two groups (A, D–G, I, K) or ordinary one-way analysis of variance (ANOVA) with Dunnett’s multiple comparisons test (C) (A, C–G, I, K; error bars represent the standard deviation [SD of biological replicates]).
-
Figure 2—source data 1
Source data for Figure 2A, C–G1, K.
- https://cdn.elifesciences.org/articles/78069/elife-78069-fig2-data1-v2.xlsx
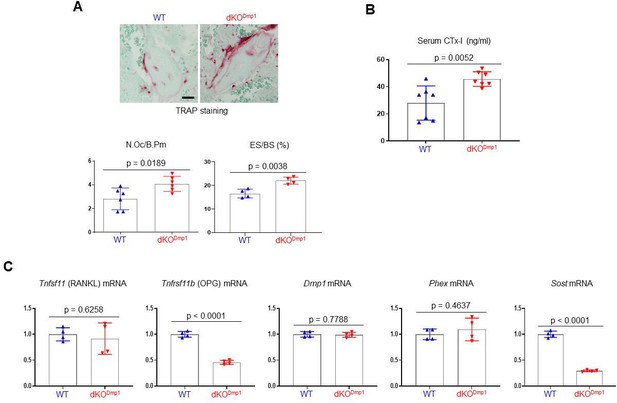
Analysis of skeletal phenotypes in dKODmp1 mice.
(A) Tartrate-resistant acid phosphatase (TRAP)-stained longitudinal sections of 8-week-old WT and dKODmp1 femurs. Representative images (top) and the relative quantification (bottom) are shown. N.Oc/B.pm, osteoclast number per bone perimeter; ES/BS, erosion surface per bone surface. Scale bar, 20 µm (top). (B) Serum levels of cross-linked C-telopeptide of type 1 collagen (CTx-I) in 8-week-old WT and dKODmp1 male mice were measured by ELISA. (C) mRNA levels of Tnfsf11, Tnfrsf11b, Dmp1, Phex, and Sost in 8-week-old WT and dKODmp1 tibial bones were assessed by RT-PCR. Data are representative of two or three independent experiments (A) [top], (C) or pooled from two experiments (A) [bottom], (B). A two-tailed unpaired Student’s t-test for comparing two groups (A–C; error bars represent the standard deviation [SD] of biological replicates).
-
Figure 2—figure supplement 1—source data 1
Source data for Figure 2—figure supplement 1A–C.
- https://cdn.elifesciences.org/articles/78069/elife-78069-fig2-figsupp1-data1-v2.xlsx
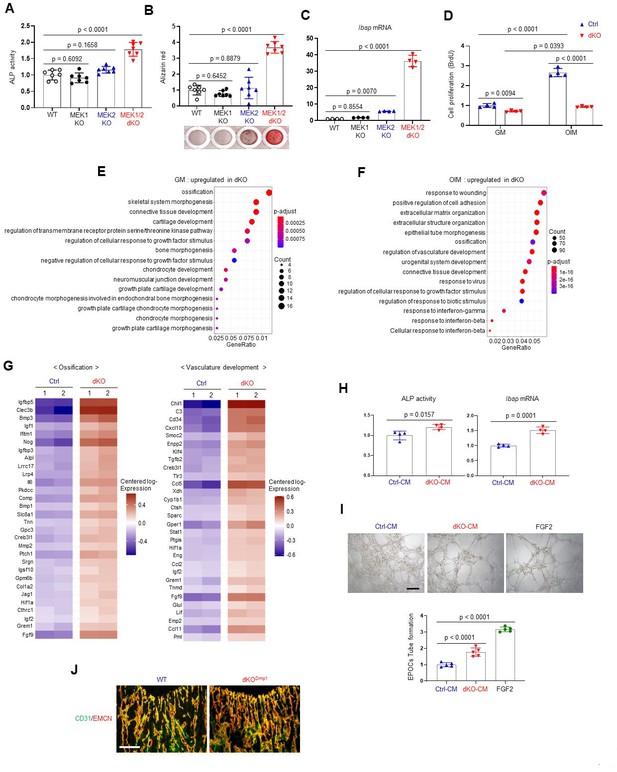
Effects of MEK1/2 deletion on osteoblast differentiation and production of angiogenic and osteogenic factors.
(A–C) Mouse Map2k1fl/fl;Map2k2+/+ and Map2k1fl/fl;Map2k2−/− osteoblasts (Obs) were infected with control vector or Cre recombinase-expressing lentiviruses; Map2k1fl/fl;Map2k2+/+ Obs with control (WT) or Cre (MEK1 KO), Map2k1fl/fl;Map2k2−/− Obs with control (MEK2 KO) or Cre (MEK1/2 dKO). Puromycin-selected Obs were cultured under osteogenic conditions and alkaline phosphatase (ALP) activity (A) and osteogenic gene expression (C) were determined at day 6 and mineralization (B) was analyzed at day 18 of culture. (D) MEK2 KO (Ctrl) and MEK1/2 dKO (dKO) Obs were cultured with control growth medium (GM) or osteogenic induction medium (OIM) and cell proliferation was analyzed by Bromodeoxyuridine (BrdU) incorporation at day 6 of the culture. (E, F) Transcriptome analysis of Ctrl and dKO Obs 6 days after GM or OIM culture. Biological process output of gene ontology analysis was performed in both GM and OIM group for upregulated genes in dKO relative to Ctrl Obs. The color indicates adjusted p value as estimated by the Benjamini–Hochberg method with the threshold of significance p = 0.05 and q = 0.005. (G) Heatmaps for ossification- and vasculature-associated gene expression. The top 30 upregulated genes in dKO Obs relative to Ctrl Obs are displayed as each row and column represent gene symbol and sample, respectively. The log10 expression (read count) was centered across samples and red and purple denote upregulated and downregulated, respectively. (H) Conditioned medium (CM) from Ctrl and dKO Obs was collected at day 6 under osteogenic culture and mouse wildtype bone marrow-derived mesenchymal stromal cells (BMSCs) were cultured under osteogenic condition in the presence of CM of Ctrl Obs and dKO Obs and ALP activity (left) and Ibsp mRNA level (right) were assessed at day 6. (I) Capillary tube formation of mouse endothelial cells (EPOCs) was performed in the presence of CM for 5 hr. Representative images (top) and quantification for the number of branches are displayed (bottom). FGF2 was used as a positive control. Scale bar, 200 µm. (J) Immunofluorescence for CD31 (green) and endomucin (EMCN, red) in the epiphyseal area of 8-week-old WT and dKODmp1 femurs. Scale bar, 100 µm. Data are representative of three independent experiments (A–D, H–J). For transcriptome analysis, biological duplicates were analyzed (E–G). Ordinary one-way analysis of variance (ANOVA) with Dunnett’s multiple comparisons test (A–C, I) or a two-tailed unpaired Student’s t-test for comparing two groups (D, H) (A–D, H, I; error bars, standard deviation [SD] of biological replicates).
-
Figure 3—source data 1
Source data for Figure 3A–D, H1.
- https://cdn.elifesciences.org/articles/78069/elife-78069-fig3-data1-v2.xlsx
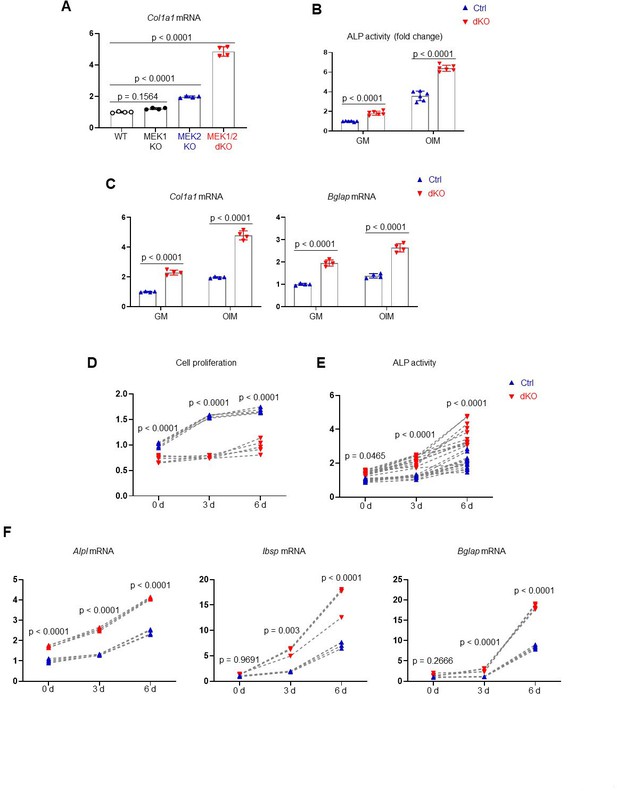
Characterization of MEK1/2-deficient osteoblast-lineage cells.
(A) Mouse Map2k1fl/fl;Map2k2+/+ and Map2k1fl/fl;Map2k2−/− osteoblasts (Obs) were infected with vector or Cre recombinase-expressing lentiviruses; Map2k1fl/fl;Map2k2+/+ Obs with vector (WT) or Cre (MEK1 KO), Map2k1fl/fl;Map2k2−/− Obs with vector (MEK2 KO) or Cre (MEK1/2 dKO). Puromycin-selected Obs were cultured under osteogenic conditions and Col1a1 gene expression was determined at day 6. (B, C) Ctrl (MEK2 KO) and dKO (MEK1/2 dKO) Obs were cultured in the presence of growth medium (GM) or osteogenic induction medium (OIM) and alkaline phosphatase (ALP) activity (B) and osteogenic gene expression (C) were assessed at day 6 in culture. Cell proliferation (D), ALP activity (E), and osteogenic gene expression (F) of Ctrl and dKO Obs at days 0, 3, and 6 of the osteogenic culture were analyzed by alamar blue staining, colorimetric assay, and RT-PCR, respectively. Data are representative of three independent experiments. An ordinary one-way analysis of variance (ANOVA) with Dunnett’s multiple comparisons test (A) or a two-tailed unpaired Student’s t-test for comparing two groups (B–F) (A–F; error bars represent the standard deviation [SD] of biological replicates).
-
Figure 3—figure supplement 1—source data 1
Source data for Figure 3—figure supplement 1A–F.
- https://cdn.elifesciences.org/articles/78069/elife-78069-fig3-figsupp1-data1-v2.xlsx
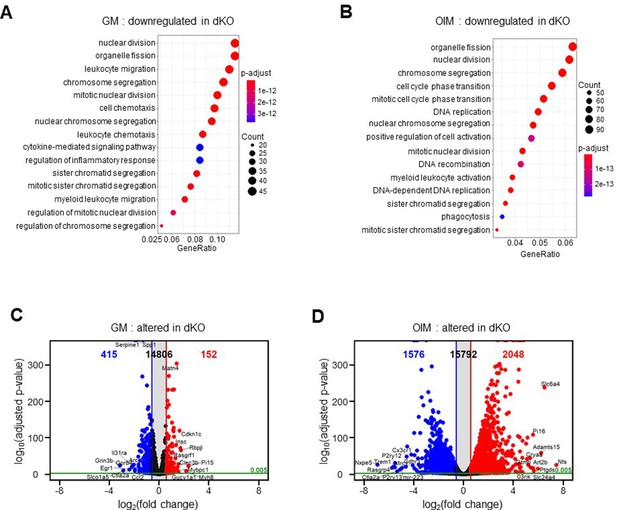
Transcriptome analysis of MEK1/2-deficient osteoblast-lineage cells.
(A–D) Transcriptome analysis of Ctrl and dKO Obs 6 days after growth medium (GM) or osteogenic induction medium (OIM) culture. Biological process output of gene ontology analysis was performed in both GM (A) and OIM (B) group for downregulated genes in dKO Obs relative to Ctrl Obs. The color indicates adjusted p value as estimated by the Benjamini–Hochberg method with the threshold of significance p = 0.05 and q = 0.005. Volcano plots showing the gene expression for up/downregulated genes in dKO Obs relative to Ctrl Obs after GM (C) or OIM (D) culture. Dots indicate upregulated (red) and downregulated genes (blue). The top 10 up/downregulated genes were labeled.
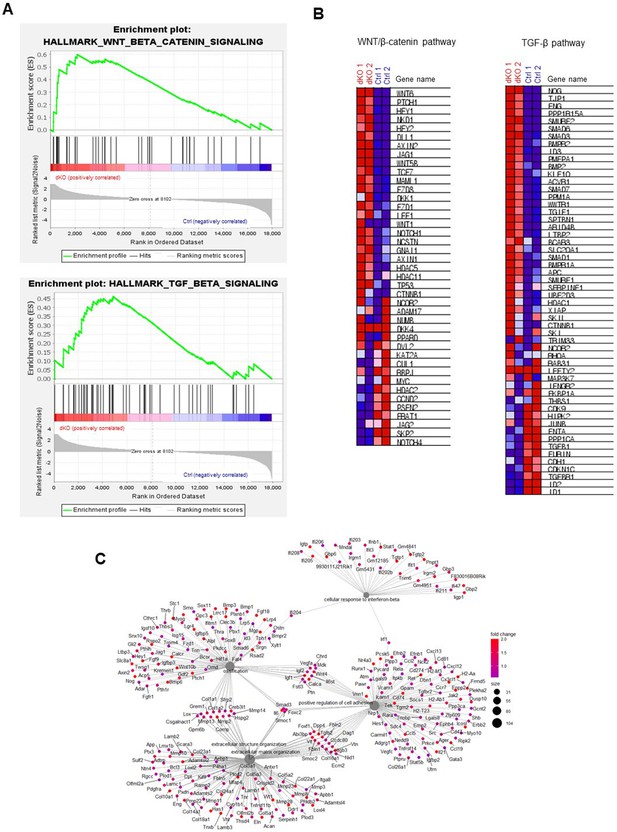
MEK1/2-deficient osteoblasts show gene enrichment of WNT and TGF-β signaling.
(A, B) Gene set enrichment analysis (GSEA) of Ctrl and dKO Obs at day 6 of the osteogenic culture shows an enrichment of genes associated with the pathways of WNT/β-catenin and TGF-β. Enrichment plots (A) and heatmap of signature gene sets (B) are displayed. (C) Category net plot of Ctrl and dKO Obs at day 6 of the osteogenic culture shows a relationship between genes associated with the significant gene ontology (GO) term.
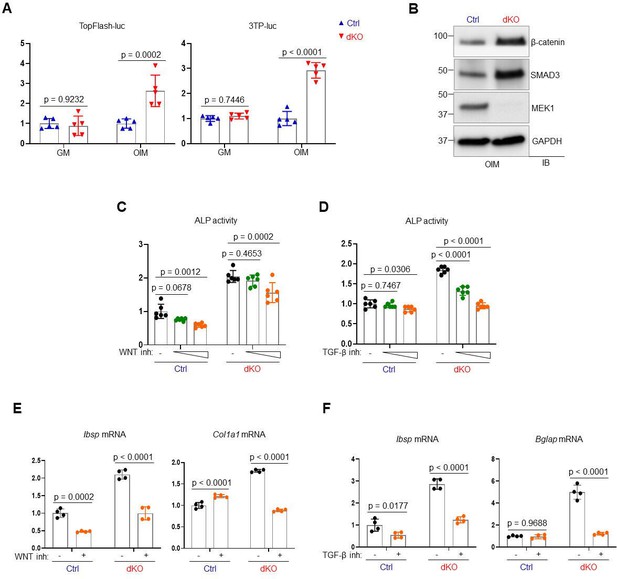
MEK1/2-deficient osteoblasts show enhanced WNT/β-catenin and TGF-β signaling at late stages of osteogenic differentiation.
(A) Ctrl and dKO Obs were cultured under growth (GM) or osteogenic conditions (OIM) for 3 days and transfected with a Topflash-luc or 3TP-luc reporter gene along with Renilla. 48 hr later, luciferase activity was measured and normalized to Renilla activity. (B) Immunoblotting analysis showing protein levels of β-catenin and SMAD3 in Ctrl and dKO Obs at day 6 of osteogenic culture (OIM). GAPDH was used as a loading control. (C–F) Ctrl and dKO Obs were cultured under osteogenic conditions in the presence of vehicle or an inhibitor against WNT or TGF-β signaling and 6 days later, alkaline phosphatase (ALP) activity (C, D) and osteogenic gene expression (E, F) were analyzed for osteogenic differentiation. A two-tailed unpaired Student’s t-test (A, E, F) or an ordinary one-way analysis of variance (ANOVA) with Dunnett’s multiple comparisons test (C, D) was used to compare groups (A, C–F; error bars represent the standard deviation [SD] of biological replicates).
-
Figure 3—figure supplement 4—source data 1
Source data for Figure 3—figure supplement 4A, C–F.
- https://cdn.elifesciences.org/articles/78069/elife-78069-fig3-figsupp4-data1-v2.xlsx
-
Figure 3—figure supplement 4—source data 2
Full immunoblots for Figure 3—figure supplement 4B.
- https://cdn.elifesciences.org/articles/78069/elife-78069-fig3-figsupp4-data2-v2.pdf
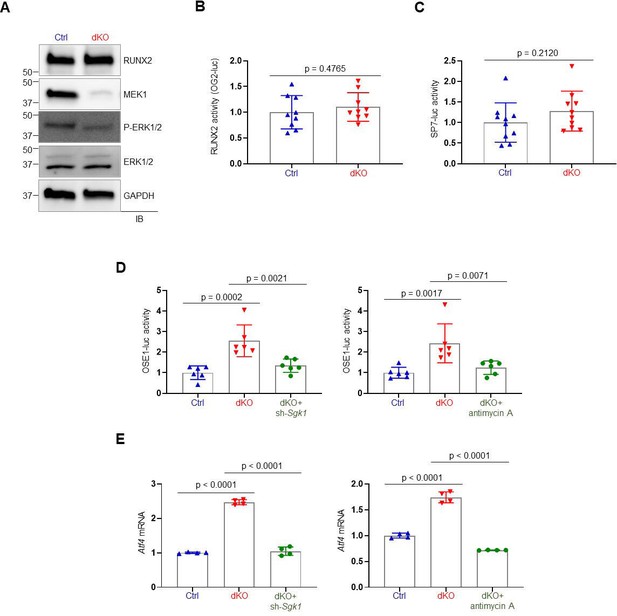
Effects of MEK1/2 deletion on RUNX2 expression and transcriptional activity in osteoblasts.
(A) Immunoblotting analysis showing protein levels of RUNX2 in Ctrl and dKO Obs. GAPDH was used as a loading control. (B, C) Ctrl and dKO Obs were transfected with the OG2-luc reporter gene or SP7-responsive reporter gene along with Renilla. After 48 hr, luciferase activity was measured and normalized to Renilla activity. (D) Ctrl and dKO Obs expressing sh-Scr or sh-Sgk1 shRNAs (left) or treated with antimycin A (mitochondria inhibitor, 10 µM, right) were transfected with the ATF4-responsive reporter gene (OSE1-luc) along with Renilla. After 48 hr, luciferase activity was measured and normalized to Renilla activity. (E) Atf4 mRNA levels of Ctrl and dKO Obs expressing sh-Scr or sh-Sgk1 shRNAs (left) or treated with antimycin A (10 µM) (right) were examined at day 6 of the osteogenic culture. Data are representative of two independent experiments. A two-tailed unpaired Student’s t-test (B, C) or an ordinary one-way analysis of variance (ANOVA) with Dunnett’s multiple comparisons test (D, E) was used to compare groups (B–E; error bars represent the standard deviation [SD] of biological replicates).
-
Figure 3—figure supplement 5—source data 1
Full immunoblots for Figure 3—figure supplement 5A.
- https://cdn.elifesciences.org/articles/78069/elife-78069-fig3-figsupp5-data1-v2.pdf
-
Figure 3—figure supplement 5—source data 2
Source data for Figure 3—figure supplement 5B–E.
- https://cdn.elifesciences.org/articles/78069/elife-78069-fig3-figsupp5-data2-v2.xlsx
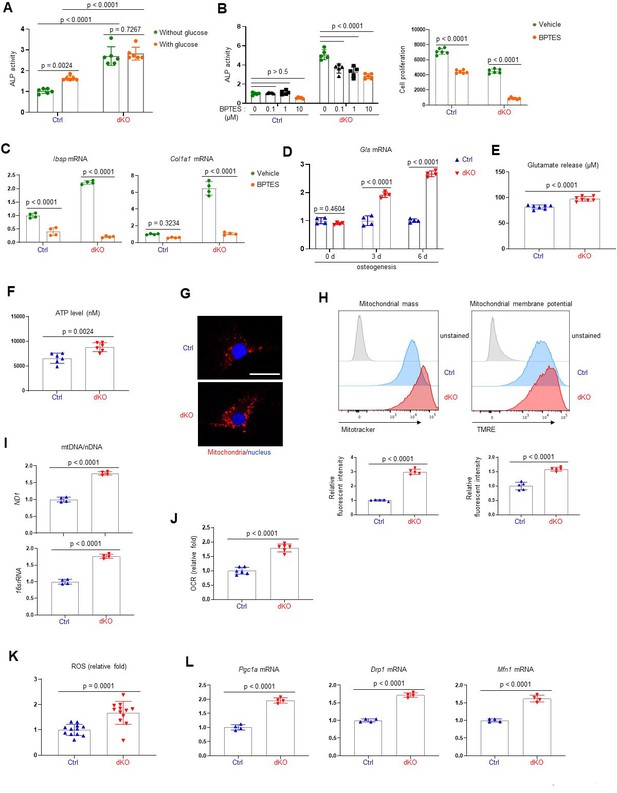
MEK1/2 deletion enhances mitochondria-mediated energy metabolism in osteoblasts.
(A) Ctrl and dKO Obs were cultured under osteogenic conditions in the presence or absence of glucose and 6 days later, alkaline phosphatase (ALP) activity was assessed. Ctrl and dKO Obs were treated with different doses of bis-2-[5-phenylacetamido-1,3,4-thiadiazol-2-yl]ethyl sulfide (BPTES) under osteogenic conditions and 6 days later, ALP activity and cell proliferation (B) and osteogenic gene expression (C) were assessed. mRNA levels of Gls in Ctrl and dKO Obs were measured by RT-PCR analysis at different time points of osteogenic culture (D) and levels of glutamate in the supernatant were measured for extracellular glutamate release at day 6 culture of osteogenesis (E). (F–I) Intracellular adenosine triphosphate (ATP) levels (F), mitochondrial numbers (G), and the ratio of mitochondrial DNA (mtDNA, mt-ND1 or mt-16sRNA) to nuclear DNA (nDNA, Hk2) (I) were assessed in Ctrl and dKO Obs after 6 days of culture. Alternatively, flow cytometry was used to measure mitochondrial numbers and membrane potential of Ctrl and dKO Obs treated with Mitotracker or TMRE at day 4 of osteogenic culture, respectively (H). Numbers indicate median fluorescence intensity (H, bottom). Scale bar, 75 µm (G). (J) Oxygen consumption rate (OCR) in Ctrl and dKO Obs 6 days after osteogenic culture. (K) Intracellular reactive oxidative species (ROS) levels in Ctrl and dKO Obs were assessed 6 days after osteogenic culture. (L) mRNA levels of mitochondria-related genes in Ctrl and dKO Obs were assessed by RT-PCR analysis. Data are representative of three independent experiments. A two-tailed unpaired Student’s t-test for comparing two groups (A, B) [right], (C–F, H–L) or ordinary one-way analysis of variance (ANOVA) with Dunnett’s multiple comparisons test (B [left]) (A–F, H–L; error bars, standard deviaiton [SD] of biological replicates).
-
Figure 4—source data 1
Source data for Figure 4A–F, H–L.
- https://cdn.elifesciences.org/articles/78069/elife-78069-fig4-data1-v2.xlsx
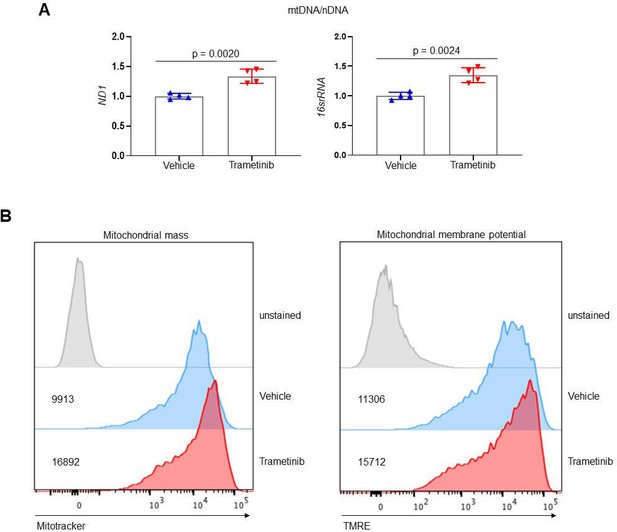
Extracellular signal-regulated kinase (ERK) inhibition enhances mitochondrial function in osteoblasts.
(A) Mouse wildtype Obs were treated with vehicle or trametinib (0.5 µM) under osteogenic conditions and 6 days later, mitochondrial DNA (mtDNA) copy number was determined by mitochondrial DNA (mtDNA, mt-ND1 or mt-16sRNA) to nuclear DNA (nDNA, Hk2) ratio using RT-PCR. (B) Mitochondrial mass (left) and membrane potential (right) in vehicle- or 0.5 μM trametinib-treated Obs were analyzed using Mitotracker and TMRE staining, respectively, at day 4 of the osteogenic culture. Numbers indicate median fluorescence intensity. Data are representative of three independent experiments. A two-tailed unpaired Student’s t-test for comparing two groups (A; error bars represent the standard deviation [SD] of biological replicates).
-
Figure 4—figure supplement 1—source data 1
Source data for Figure 4—figure supplement 1A.
- https://cdn.elifesciences.org/articles/78069/elife-78069-fig4-figsupp1-data1-v2.xlsx
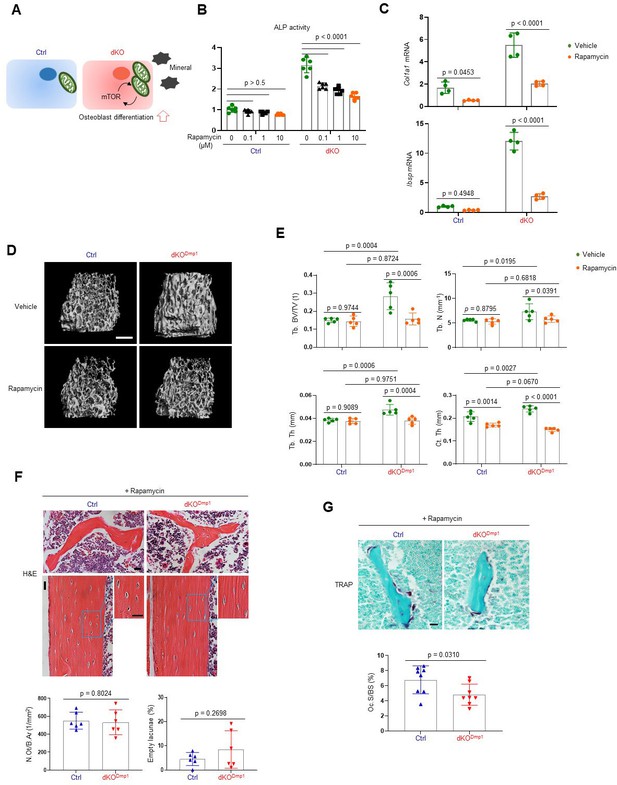
Rapamycin treatment reverses skeletal phenotypes of MEK1/2-deficient mice.
(A) Diagram showing the mechanisms by which extracellular signal-regulated kinase (ERK) inhibition enhances osteoblast differentiation due to augmented mechanistic target of rapamycin (mTOR)-mediated mitochondrial function. Ctrl and dKO Obs were treated with different doses of rapamycin under osteogenic conditions and 6 days later, alkaline phosphatase (ALP) activity (B) and osteogenic gene expression (C) were determined. (D, E) MicroCT analysis showing femoral bone mass in 8-week-old Ctrl and dKODmp1 mice treated with vehicle or rapamycin. 3D reconstruction (D) and the relative quantification (E) are displayed. Scale bar, 500 µm (D). (F) Hematoxylin and eosin (H&E)-stained longitudinal sections of 8-week-old Ctrl and dKODmp1 femurs treated with rapamycin. Representative images (top) and numbers of osteocytes/bone area (N.Ot/B.Ar) and empty lacunae (bottom) are displayed. Scale bar, 20 µm (top). (G) Tartrate-resistant acid phosphatase (TRAP)-stained longitudinal sections of 8-week-old Ctrl and dKODmp1 femurs treated with rapamycin. Representative images (top) and osteoclast surface/bone surface (Oc.S/BS) (bottom) are displayed. Scale bar, 20 µm (top). Data are representative of two or three independent experiments (B–D, F [top], G [top]) or pooled from two experiments (E, F [bottom], G [bottom]). Ordinary one-way analysis of variance (ANOVA) with Dunnett’s multiple comparisons test (B) or a two-tailed unpaired Student’s t-test for comparing two groups (C, E–G; B, C, E–G; error bars, standard deviation [SD] of biological replicates).
-
Figure 5—source data 1
Source data for Figure 5B, C, E–G.
- https://cdn.elifesciences.org/articles/78069/elife-78069-fig5-data1-v2.xlsx

Rapamycin inhibits both mTORC1 and mTORC2 pathways.
Immunoblotting analysis showing protein levels of P-AKT/AKT, P-SGK1/SGK1, and P-p70S6K/p70S6K in dKO Obs. GAPDH was used as a loading control. Data are representative of three independent experiments.
-
Figure 5—figure supplement 1—source data 1
Full immunoblots for Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/78069/elife-78069-fig5-figsupp1-data1-v2.pdf
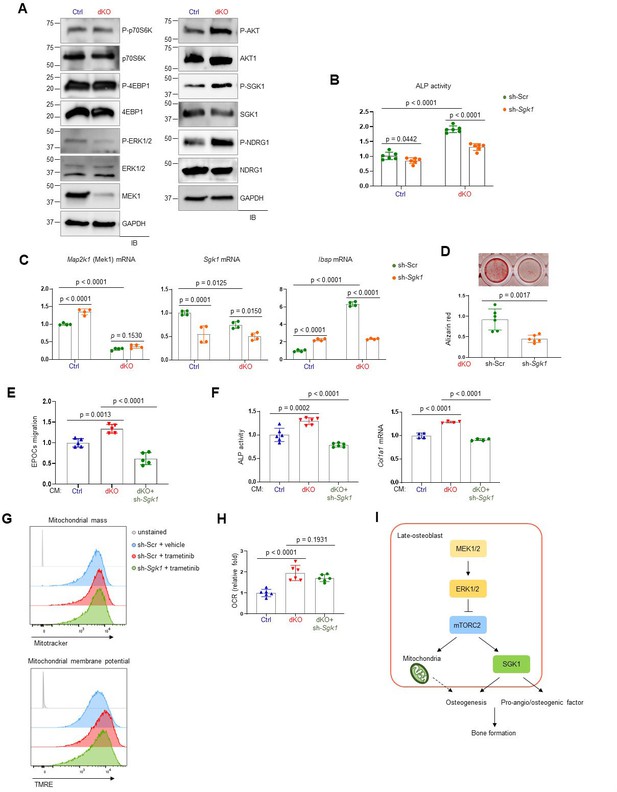
mTORC2/SGK1 activation is important for MEK1/2 deficiency-induced osteoblast differentiation.
(A) Immunoblot analysis showing phosphorylation levels of mechanistic target of rapamycin (mTOR) signaling components in Ctrl and dKO Obs. GAPDH was used as a loading control. (B–D) Ctrl and dKO Obs expressing sh-Scr or sh-Sgk1 shRNAs were cultured under osteogenic conditions. Alkaline phosphatase (ALP) activity (B), osteogenic markers expression (C), and mineralization (D) were assessed after 6 or 18 days of culture, respectively. (E) Conditioned medium (CM) collected from Ctrl and dKO Obs expressing sh-Scr or sh-Sgk1 shRNAs 6 days after osteogenic culture was added to transwell migration of mouse endothelial cells (EPOCs) and migrated cells were assessed 12 hr after incubation. (F) Mouse bone marrow-derived mesenchymal stromal cells (BMSCs) were cultured under osteogenic condition in the presence of CM, and ALP activity and Cola1 mRNA level were analyzed at day 6 of the culture. (G) Vehicle- or trametinib-treated Obs expressing sh-Scr or sh-Sgk1 shRNAs were treated with Mitotracker or TMRE 4 days after osteogenic culture and mitochondrial mass and membrane potential were assessed using flow cytometry, respectively. (H) Oxygen consumption rate (OCR) in Ctrl and dKO Obs expressing sh-Scr or sh-Sgk1 shRNAs after 6 days of culture. (I) Diagram showing the molecular actions of extracellular signal-regulated kinase (ERK) on bone formation. Data are representative of two or three independent experiments (A–H). A two-tailed unpaired Student’s t-test for comparing two groups (B–D) or ordinary one-way analysis of variance (ANOVA) with Dunnett’s multiple comparisons test (E, F, H; B–F, H; error bars, standard deviation [SD] of biological replicates).
-
Figure 6—source data 1
Full immunoblots for Figure 6A.
- https://cdn.elifesciences.org/articles/78069/elife-78069-fig6-data1-v2.pdf
-
Figure 6—source data 2
Source data for Figure 6B–F, H.
- https://cdn.elifesciences.org/articles/78069/elife-78069-fig6-data2-v2.xlsx
Tables
Primer sequences.
| Gene | Forward | Reverse |
|---|---|---|
| Human RUNX2 | TTACTTACACCCCGCCAGTC | CACTCTGGCTTTGGGAAGAG |
| Human SP7 | TTACAAGCACTAATGGGCTCCT | GTAGACACTGGGCAGACAGTCA |
| Human RPLP0 | GGAATGTGGGCTTTGTGTTC | TGCCCCTGGAGATTTTAGTG |
| Mouse Alpl | CACAATATCAAGGATATCGACGTGA | ACATCAGTTCTGTTCTTCGGGTACA |
| Mouse Ibsp | CAGGGAGGCAGTGACTCTTC | AGTGTGGAAAGTGTGGCGTT |
| Mouse Bglap | GCAGCACAGGTCCTAAATAG | GGGCAATAAGGTAGTGAACAG |
| Mouse Col1a1 | ACTGTCCCAACCCCCAAAG | ACGTATTCTTCCGGGCAGAA |
| Mouse Atf4 | CCACTCCAGAGCATTCCTTTAG | TGTCATTGTCAGAGGGAGTGTC |
| Mouse Map2k1 (MEK1) | ACTGCCCAGTGGAGTATTCAGT | TGTACCATGAGCTGCTTCAGAT |
| Mouse Map2k2 (MEK2) | CATCAGTGTAGGTCATGGGATG | AAACTCCTGGAAGTCTGAGCTG |
| Mouse Tnfsf11 (RANKL) | CAGCATCGCTCTGTTCCTGTA | CTGCGTTTTCATGGAGTCTCA |
| Mouse Tnfrsf11b (OPG) | CGGAAACAGAGAAGCCACGCAA | CTGTCCACCAAAACACTCAGCC |
| Mouse Dmp1 | GAAAGCTCTGAAGAGAGGACGG | CCTCTCCAGATTCACTGCTGTC |
| Mouse Phex | CTGGCTGTAAGGGAAGACTCCC | GCTCCTAAAAGCACAGCAGTGTC |
| Mouse Sost | CTTCAGGAATGATGCCACAGAGGT | ATCTTTGGCGTCATAGGGATGGTG |
| Mouse Gls | GGCAAAGGCATTCTATTGGA | CTTGGCTCCTTCCCAACATA |
| Mouse Pgc1a (Ppargc1a) | CCCACAACTCCTCCTCATAAAG | TTGGGTACCAGAACACTCACTG |
| Mouse Drp1 | GGGGTAAATTTCTTCACACCAA | TCAGGGCTTACCCCCTTATTAT |
| Mouse Mfn1 | ATAGAAGATGGCATGGGAAGAA | GCTGGAAGTAGTGGCTTCAAGT |
| Mouse Sgk1 | GGGTGCCAAGGATGACTTTA | TGGGTTAAATGGGGGTGTAA |
| Mouse Rplp0 | TGGCCAATAAGGTGCCAGCTGCTG | CTTGTCTCCAGTCTTTATCAGCTGCAC |
| Mouse Hk2 | GCCAGCCTCTCCTGATTTTAGTGT | GGGAACACAAAAGACCTCTTCTGG |
| mt-ND1 | CTAGCAGAAACAAACCGGGC | CCGGCTGCGTATTCTACGTT |
| mt-16srRNA | CCGCAAGGGAAAGATGAAAGAC | TCGTTTGGTTTCGGGGTTTC |





