Process- and product-related impurities in the ChAdOx1 nCov-19 vaccine
Figures
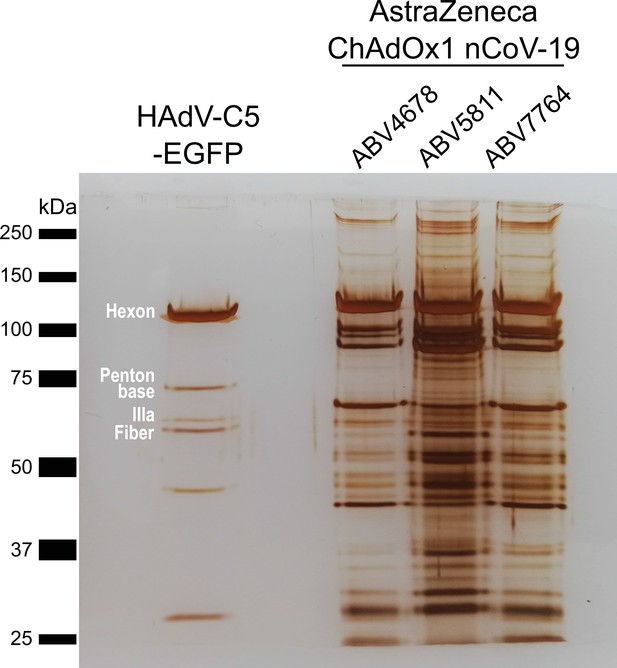
Protein staining of HAdV-C5-EGFP and three ChAdOx1 nCoV-19 vaccine lots.
3 × 109 adenoviral vector particles were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) under denaturing and reducing conditions. Proteins were visualized by silver staining. Known HAdV-C5 proteins are labeled. Three different vaccine lots (ABV4678, ABV5811, and ABV7764) of ChAdOx1, produced by the manufacturer, were analyzed.
-
Figure 1—source data 1
Original file of the full raw unedited gel: Protein staining of HAdV-C5-EGFP and three ChAdOx1 nCov-19 vaccine lots.
- https://cdn.elifesciences.org/articles/78513/elife-78513-fig1-data1-v2.pdf
-
Figure 1—source data 2
Uncropped gel with the relevant bands labeled: Protein staining of HAdV-C5-EGFP and three ChAdOx1 nCov-19 vaccine lots.
- https://cdn.elifesciences.org/articles/78513/elife-78513-fig1-data2-v2.pdf
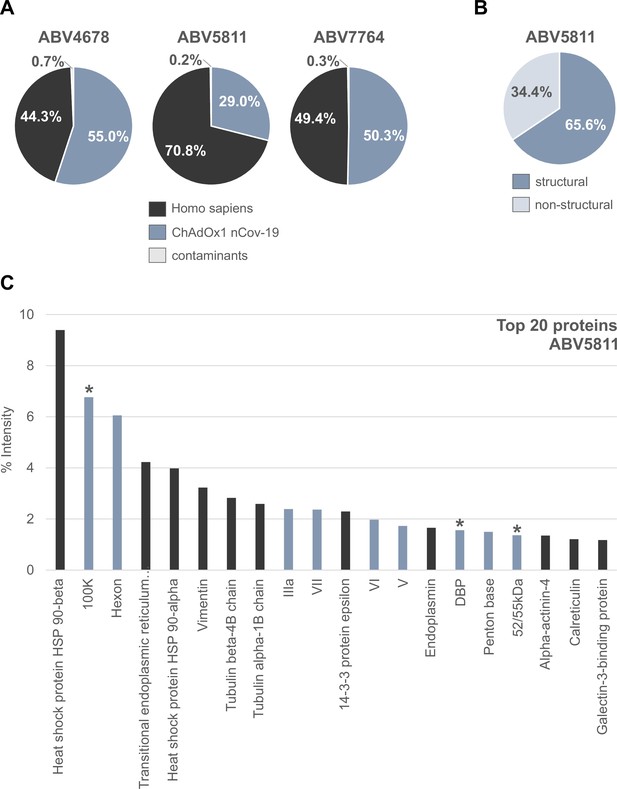
Distribution of proteins in three ChAdOx1 nCov-19 vaccine lots.
The protein composition of three lots of ChAdOx1 (ABV4678, ABV5811, and ABV7764) was analyzed by mass spectrometry following in-solution protein digest. Spectral data were aligned via search engine with human and viral databases (Figure 2—source data 1). (A) Percentage of total intensities associated with proteins from the respective organism were subsequently summed. (B) Percentage of structural and nonstructural adenoviral proteins detected in ChAdOx1 nCov-19 vaccine lot ABV5811. (C) Intensity distribution of the top 20 proteins of the ChAdOx1 nCov-19 vaccine lot ABV5811 detected. Proteins that originate from Homo sapiens are depicted in black; proteins that originate from ChAdOx1 are depicted in blue-gray; *nonstructural adenoviral proteins.
-
Figure 2—source data 1
List of proteins detected in three ChAdOx1 nCov-19 vaccine lots by mass spectrometry.
- https://cdn.elifesciences.org/articles/78513/elife-78513-fig2-data1-v2.xlsx
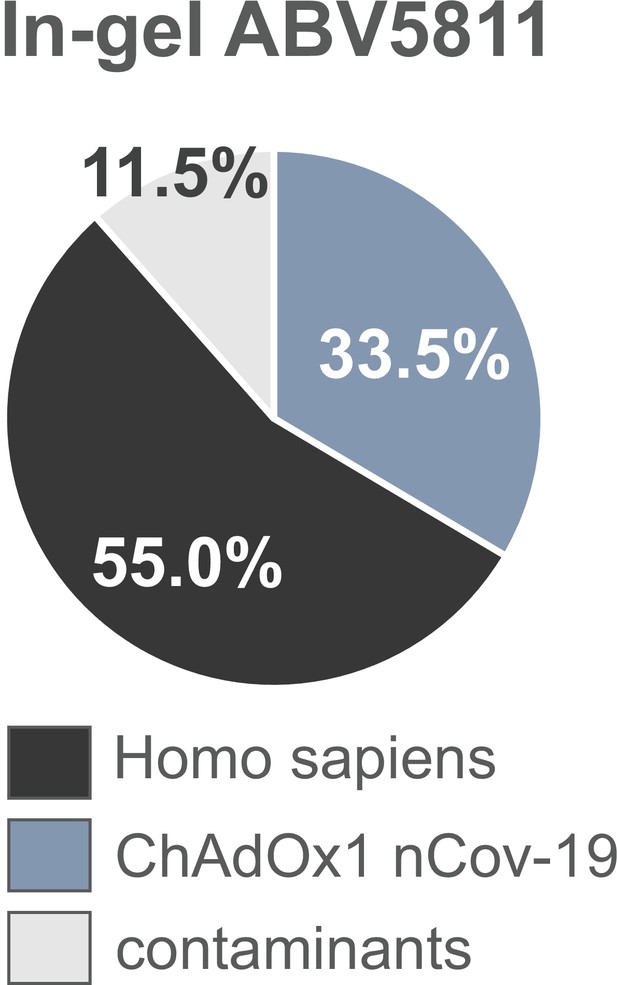
Distribution of proteins of the ChAdOx1 nCov-19 vaccine following PAGE and in-gel digests.
The protein composition of lot ABV5811 was analyzed by mass spectrometry following in-gel protein digests. Spectral data were aligned via search engine with human and viral databases. Percentage of total intensities associated with proteins from the respective organism were subsequently summed (Figure 2—source data 1).
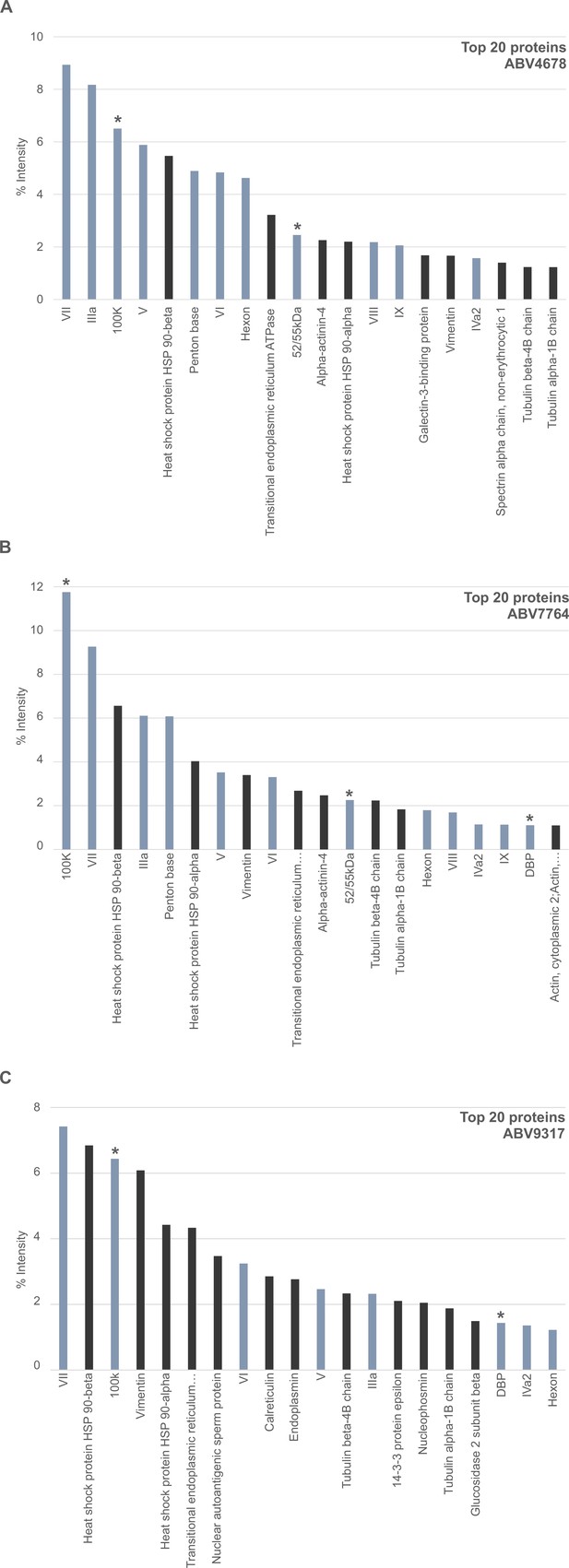
Intensity distribution of the top 20 proteins in ChAdOx1 nCov-19 vaccine lots.
The protein composition of ChAdOx1 nCov-19 vaccine lots was analyzed by mass spectrometry following in-solution protein digest. Spectral data were aligned via search engine with human and viral databases and intensity distribution of the top 20 proteins are shown. Proteins that originate from Homo sapiens are depicted in black; proteins that originate from ChAdOx1 are depicted in blue-gray; *nonstructural adenoviral proteins. (A) ABV4678, (B) ABV7764, and (C) ABV9317 (Figure 2—source data 1).
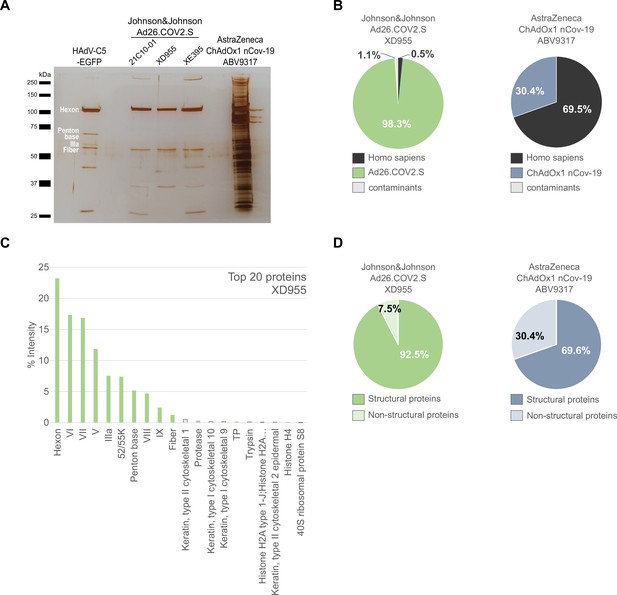
Comparison of Ad26.COV2.S and ChAdOx1 nCov-19 vaccines by biochemical and proteomic analysis.
(A) Proteins corresponding to each 3 × 109 vector particles of three lots of Ad26.COV2.S (21C10-01, XD955, and XE395), one lot of ChAdOx1 (ABV9317) and CsCl-purified HAdV-C5-EGFP (control), as indicated, were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) performed under denaturing and reducing conditions. Proteins were visualized by silver staining. Known HAdV-C5 proteins are indicated. Note that on SDS–PAGE the stained protein band corresponding to Penton Base of HAdV-C5 (571 amino acids, predicted molecular weight [MW]: 63.4 kDa) has a slower migration behavior than expected, likely due to a flexible loop that is structurally disordered in HAdV-C5 (Flatt et al., 2013) and known to be hypervariable in different adenovirus types (Zubieta et al., 2005). Compared to HAdV-C5, in Chimpanzee Adenovirus Y25 (532 amino acids, MW: 532 kDa) and in HAdV-D26 (519 amino acids, MW: 58.6) this loop is reduced in size and, according to the PONDR-Fit online tool (Xue et al., 2010), less disordered (data not shown), explaining, why in Ad26.COV2.S and in ChAdOx1 the Penton Base-corresponding signals likely overlap with those of the smaller viral proteins IIIa or fiber. (B–D) Ad26.COV2.S (lot XD955) and ChAdOx1 nCov-19 (lot ABV9317) were analyzed by mass spectrometry following in-solution protein digest. Spectral data were aligned via search engine with human and viral proteins (Figure 3—source data 1). Human proteins are depicted in black; proteins that originate from ChAdOx1 are depicted in blue-gray; proteins that originate from Ad26.COV2.S are depicted in green. (B) Percentage of total intensities of proteins from the respective organism detected in the Ad26.COV2.S (lot XD955) and ChAdOx1 nCov-19 (lot ABV9317) vaccines. (C) Intensity distribution of the top 20 proteins of the Ad26.COV2.S vaccine (lot XD955). (D) Percentage of total intensities of structural and nonstructural adenoviral proteins detected in Ad26.COV2.S (lot XD955) and ChAdOx1 nCov-19 (lot ABV9317).
-
Figure 3—source data 1
List of proteins detected by mass spectrometry in three Ad26.COV2.S vaccine lots, ChAdOx1 nCov-19 vaccine lot ABV9317, and HAdV-C5-EGFP.
- https://cdn.elifesciences.org/articles/78513/elife-78513-fig3-data1-v2.xlsx
-
Figure 3—source data 2
Original file of the full raw unedited gel: Comparison of Ad26.COV2.S and ChAdOx1 nCov-19 vaccines.
- https://cdn.elifesciences.org/articles/78513/elife-78513-fig3-data2-v2.pdf
-
Figure 3—source data 3
Uncropped gel with the relevant bands labeled: Protein staining of HAdV-C5-EGFP, three Johnson&Johnson Ad25-COV2.S vaccine lots and one AstraZeneca ChAdOx1 nCov-19 vaccine lot.
- https://cdn.elifesciences.org/articles/78513/elife-78513-fig3-data3-v2.pdf
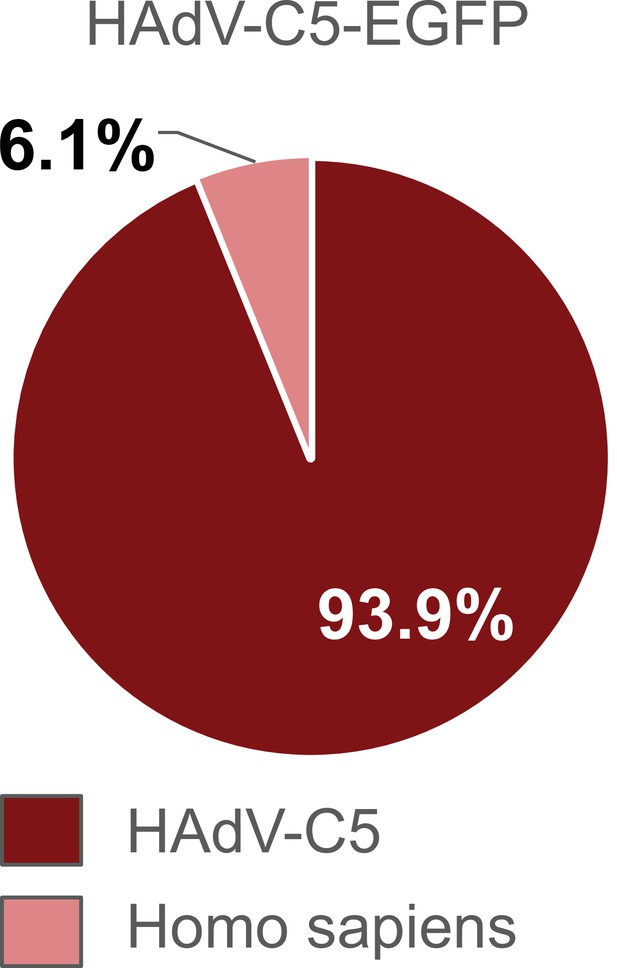
Distribution of proteins of the HAdV-C5-EGFP vector following in-solution protein digest.
HAdV-C5-EGFP was analyzed by mass spectrometry following in-solution protein digest. Spectral data were aligned via search engine with human and viral databases. Percentage of total intensities associated with proteins from the respective organism were subsequently summed. Sample processing-related contaminants were excluded in the graph (Figure 3—source data 1).
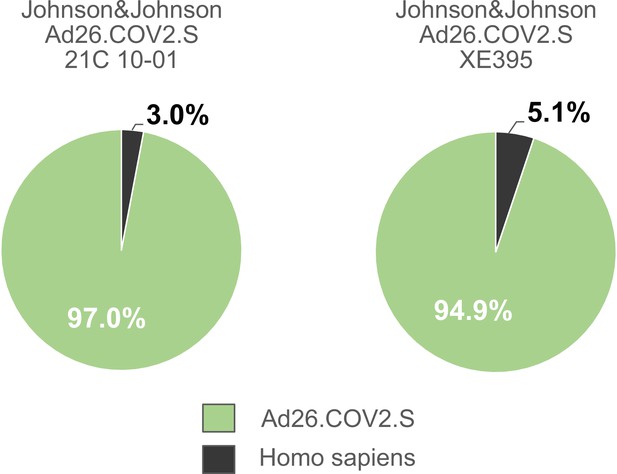
Distribution of proteins of the Johnson & Johnson Ad26.COV2.S vaccine lots 21C10-01 and lot XE395 following in-solution protein digest.
Ad26.COV2.S (lot 21C10-01 and lot XE395) were analyzed by mass spectrometry following in-solution protein digest. Spectral data were aligned via search engine with human and viral databases. Percentage of total intensities associated with proteins from the respective organism were subsequently summed. Sample processing-related contaminants were excluded in the graph (Figure 3—source data 1).
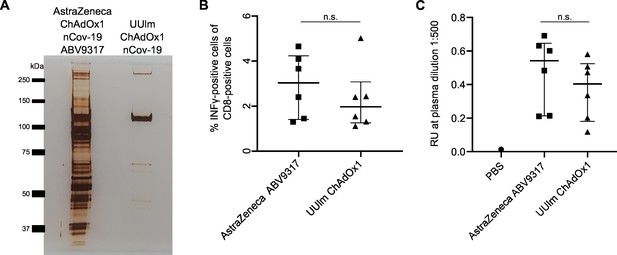
Analysis of effects of impurities in the ChAdOx1 nCoV-19 vaccine on anti-Spike immune responses in vivo.
BALB/c mice were injected intramuscularly with phosphate-buffered saline (PBS) or 1 × 109 vector particles of AstraZeneca ChAdOx1 nCoV-19 vaccine (ABV9317) or UUlm ChAdOx1 nCov-19 dissolved in PBS. Fourteen days later, mice were sacrificed and plasma and spleen samples collected. n = 6/group. (A) 3 × 109 adenoviral vector particles of AstraZeneca ChAdOx1 nCoV-19 vaccine (ABV9317) or UUlm ChAdOx1 were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) under denaturing and reducing conditions. Proteins were visualized by silver staining. (B) SARS-CoV-2 spike protein-directed T-cell responses were measured by stimulation of isolated T cells with spike peptide-loaded dimer CoVKd-268 GYLQPRTFL in duplicates and subsequent intracellular staining of INFγ. Results are given as mean % INFγ-positive cells of CD8-positive cells of single animals. (C) SARS-CoV-2 spike protein-specific antibody titers were determined by enzyme-linked immunosorbent assay (ELISA). Purified full-length spike protein was coated and serial plasma dilutions were added in duplicates. Results are given as mean response units (RU) of single animals at representative plasma dilutions of 1:500. n.s.: statistically not significant.
-
Figure 4—source data 1
List of proteins detected by mass spectrometry in UUlm ChAdOx1 nCov-19.
- https://cdn.elifesciences.org/articles/78513/elife-78513-fig4-data1-v2.xlsx
-
Figure 4—source data 2
Original file of the full raw unedited gel: Protein staining of ChAdOx1 nCov-19 vaccine lot and UUlm ChAdOx1.
- https://cdn.elifesciences.org/articles/78513/elife-78513-fig4-data2-v2.pdf
-
Figure 4—source data 3
Uncropped gel with the relevant bands labeled: Protein staining of ChAdOx1 nCoV-19 vaccine lot and UUlm ChAdOx1.
- https://cdn.elifesciences.org/articles/78513/elife-78513-fig4-data3-v2.pdf
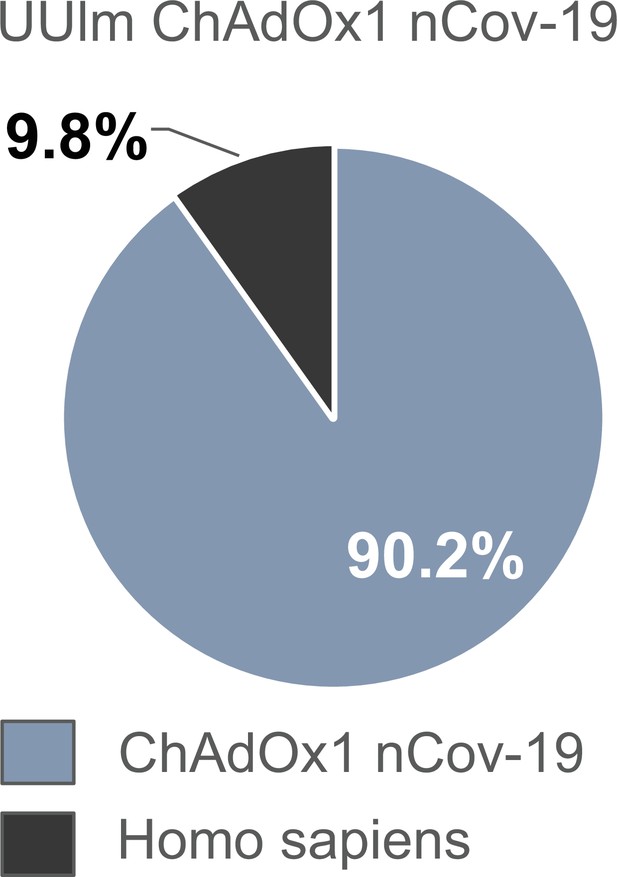
Distribution of proteins of the UUlm ChAdOx1 nCov-19 vector following in-solution protein digest.
UUlm ChAdOx1 nCov-19 was analyzed by mass spectrometry following in-solution protein digest. Spectral data were aligned via search engine with human and viral databases. Percentage of total intensities associated with proteins from the respective organism were subsequently summed. Sample processing-related contaminants were excluded in the graph (Figure 4—source data 1).
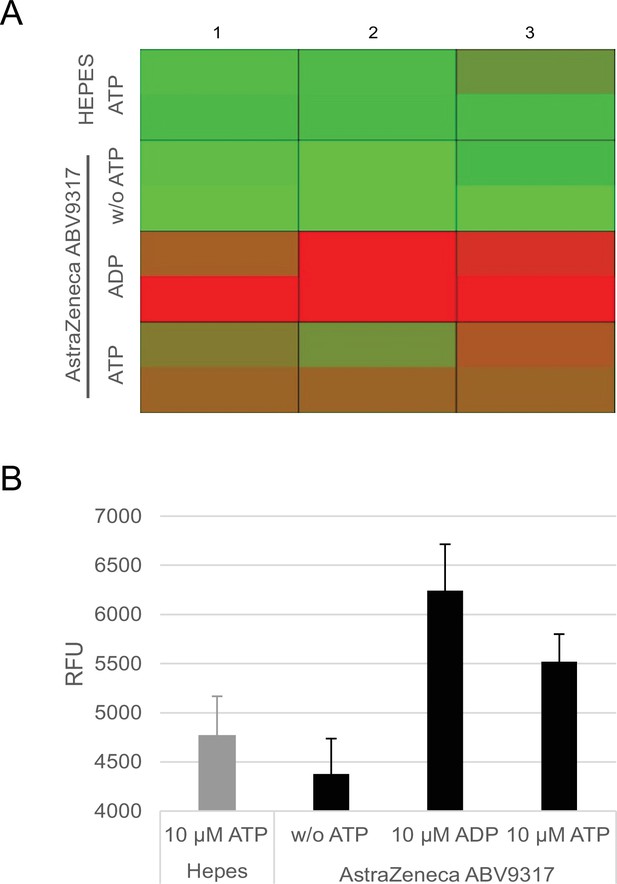
Analysis of ATPase activity in the ChAdOx1 nCoV-19 vaccine.
Conversion of ATP into ADP by ATPase activity of ChAdOx1 nCoV-19 vaccine lot ABV9317 was analyzed using a fluorescence-based commercially available ADP2 FI Assay kit, showing increasing fluorescence intensity signals with increasing amounts of ADP. Negative controls: 4-[2-hydroxyethyl]-1-piperazineethanesulfonic acid (HEPES) instead of ChAdOx1 nCov-19, no addition of ATP; Positive control: addition of ADP instead of ATP. (A) Heatmap of fluorescence intensity of Alexa594 fluorophore. (B) Quantification of measured fluorescence intensity. RFU = response fluorescence units. n = 3.
Tables
Amount of protein impurities per vaccine dose in the ChAdOx1 nCoV-19 vaccine.
European Medicines Agency (EMA) quality specifications of accepted host cell protein (HCP) amounts in the ChAdOx1 nCoV-19 vaccine and batch-release data by the manufacturer, obtained through the German information freedom act. Total protein content per dose as determined by three different methods of lots ABV4678, ABV5811, ABV7764, and ABV9317 after subtraction of 12.8 µg virus protein contained in 5 × 1010 vector particles Sweeney and Hennessey, 2002. n = 3.
| ChAdOx1 nCov-19lot # | EMA specification[µg HCP/dose] | Batch-release data manufacturer[µg HCP/dose] | NanoDrop absorbance 280 nm[µg HCP/dose] | Bradford assay[µg HCP/dose] | ImageJ analysis[µg HCP/dose] |
|---|---|---|---|---|---|
| ABV4678 | ≤0.4 | 0.05 | 39.03 | 12.36 | 9.85 |
| ABV5811 | ≤0.4 | 0.3 | 114.53 | 24.78 | 24.9 |
| ABV7764 | ≤0.4 | 0.0489 | 74.37 | 29.66 | 12.81 |
| ABV9317 | ≤0.4 | n.a. | 154.53 | 42.48 | 73.16 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Other | Covid-19 vaccine ChAdOx1 nCov-19 | AstraZeneca, distributed by the Pharmacy of the University Hospital Ulm, Germany | Lot: ABV4678 ABV5811 ABV7764 ABV9317 | Commercial vaccine |
| Other | Covid-19 vaccine Ad26-COV2.S | Johnson & Johnson, distributed by the Pharmacy of the University Hospital Ulm, Germany | Lot: XD955 21C10-01 XE395 | Commercial vaccine |
| Biological sample (Human Adenovirus type 5) | HAdV-C5-EGFP (ΔE1; CMV-eGFP) | GenBank: AY339865.1 (Δ nt 441–3522) | +CMV promoter-controlled eGFP expression cassette | |
| Cell line (Homo sapiens) | N52.E6 | Schiedner et al., 2000, Human Gene Therapy 11, 2105–2116, 2000 | ||
| Cell line (Homo sapiens) | HEK293T | ATCC | CRL-3216 | |
| Cell line (Homo sapiens) | CAP-T | Fischer et al., 2012, Biotechnology and bioengineering 109, 2250–2261, 2012 | ||
| Cell line (Rattus norvegicus, Mus musculus) | 2.4G2 | ATCC | HB-197 | Hybridoma cell line, Rattus norvegicus (B-cell), Mus muculus (myeloma) |
| Other | BALB/c mouse | Charles River | Mouse; Mus musculus; Strain code: 028 | 10–12 weeks old; n = 6/group (4× female, 2× male) |
| Peptide, recombinant protein | SARS-CoV-2 spike peptide-loaded dimer | JPT Peptide Technologies GmbH | CoVKd-268 GYLQPRTFL | |
| Recombinant DNA reagent | pBSK-CMV-Spike | This paper | Based on GenBank sequence YP_009724390.1 | CMV promoter-driven SARS-CoV-2 spike protein with some modifications |
| Antibody | Rat anti- murine-INFγ-PE, monoclonal | Invitrogen | 12-7311-82 | IF 1:200 |
| Antibody | Rat anti-murine-CD8-APC, monoclonal | eBioScience | 17-0081-83 | IF 1:200 |
| Antibody | Rabbit anti-mouse IgG, HRP-labeled, polyclonal | Sigma | A9044 | ELISA: 1:10,000 |
| Antibody | Rat/mouse anti-Fc gamma Receptor (FcRII, CD32), monoclonal | Derived from hybridoma 2.4G2 cell line (ATCC HB-197) | Four-day-old cell culture medium, undiluted | |
| Chemical compound, drug | Brefeldin A | eBioScience | 00-4506-51 | |
| Chemical compound, drug | Ni-NTA agarose beads | Qiagen | 30,210 | |
| Sequence-based reagent | Adenoviral E4 forward | GeneArt Sequences | PCR primer | TAGACGATCCCTACTGTACG |
| Sequence-based reagent | Adenoviral E4 reverse | GeneArt Sequences | PCR primer | GGAAATATGACTACGTCCGG |
| Sequence-based reagent | Adenoviral human actin forward | GeneArt Sequences | PCR primer | GCTCCTCCTGAGCGCAAG |
| Sequence-based reagent | Adenoviral human actin reverse | GeneArt Sequences | PCR primer | CATCTGCTGGAAGGTGGACA |
| Sequence-based reagent | Adenoviral human ribosomal protein L4 forward | GeneArt Sequences | PCR primer | ACGATACGCCATCTGTTCTGCC |
| Sequence-based reagent | Adenoviral human ribosomal protein L4 reverse | GeneArt Sequences | PCR primer | GGAGCAAAACAGCTTCCTTGGTC |
| Chemical compound, drug | SYBR Green | Kapa Biosystems | KK4502 | |
| Commercial assay or kit | ADP FI Assay | Bos et al., 2020, Bellbrooks Labs | 3013A | |
| Commercial assay or kit | GenElute Mammalian Genomic DNA Miniprep Kit | Sigma | G1N350 | |
| Commercial assay or kit | VenorGeM Classic | Minerva Biolabs | 11-1025 | |
| Software, algorithm | RStudio | RStudio | Version 4.0.0 | Statistics |
| Software, algorithm | MaxQuant (with Andromeda search engine; UniProt human reference proteome) | Cox and Mann, 2008; Cox et al., 2011, MaxQuant, Andromeda,UniProt | Version 1.6.3.4 | Protein identification Mass Spectrometry |





