Monoallelic CRMP1 gene variants cause neurodevelopmental disorder
Figures
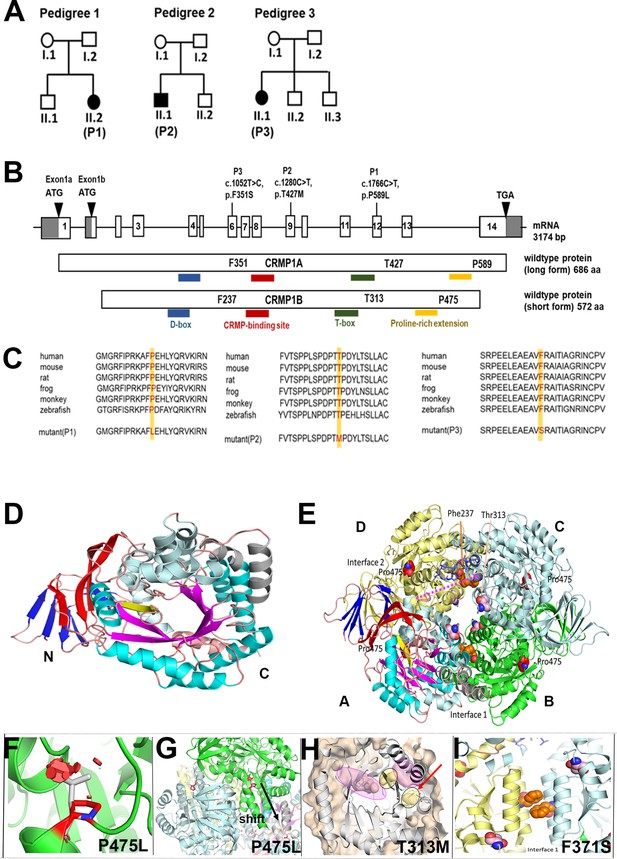
Genotype of patients with variants in CRMP1.
(A) Pedigree of index families. (B) Pictogram representing the CRMP1 cDNA with identified variant of proband 1 (P1) in exon 12 (c.1766C>T, NM_001014809.2) which leads on protein level to an amino acid change of proline to leucine in CRMP1 (CRMP1A-long form (p.P589L, NP_001014809.1) and CRMP1B-short form (p.P475L, NP_001304.1)); the variant in proband 2 (P2) in exon 9 c.280C>T (NM_001014809.2) leads to an exchange of threonine to methionine (CRMP1A (p.T427M, NP_001014809.1) and CRMP1B (p.T313M, NP_001304.1)); the variant in proband 3 (P3) in exon 6 c.052T>C (NM_001014809.2) leads to an exchange of phenyl alanine to serine (CRMP1A (p.(F351S), NP_001014809.1) and CRMP1B (p.(F237S), NP_001304.1)). (C) Multispecies sequence alignment localizes the variants in the highly conserved area of CRMP1. (D) The short form CRMP1 monomer is composed of three structural parts, an N-terminally located seven β-strands forming two β-sheets (depicted in blue), followed by a linker β-strand (yellow) connecting to the central α/β-barrel (cyan/magenta) formed by seven repeats. Inserted after repeat 4 are 2 additional α-helices (gray). (E) CRMP1 assembles into tetramers. The relevant sites of T313M, P475L, and F237S are indicated as sphere model, with the T313 and F237 located in the central channel in the vicinity of the interaction sites and P475 is located at the beginning of the C-terminal helix and oriented toward the adjacent molecules. (F) The variant P475L reveals serious clashes with neighboring residues (red hexagonals) which may be accounted for by a shift of the helix as shown in (G). (H) Detailed representation of the structural vicinity of the T313M (yellow) to the ligand-binding cavity (magenta). (I) Magnified view of interface 1 tilted 90° backwards with respect to panel E highlighting the arrangement of the two phenylalanines at position 237 from the neighboring units. The exchange of phenylalanine with hydrophobic residues to serine with hydrophilic side chain interferes with the stability of the interaction in interface 1.
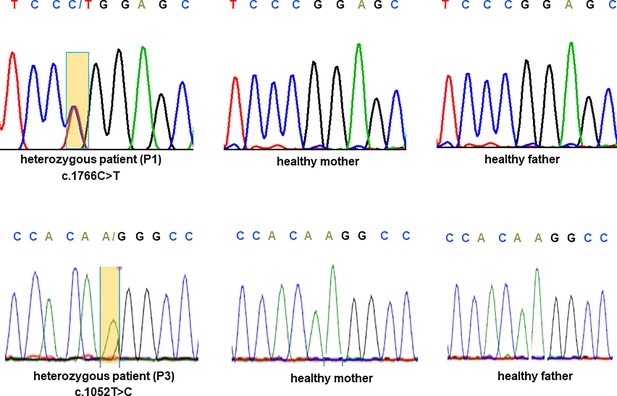
Sanger sequencing of CRMP1 in pedigrees 1 and 3.
Electropherogram traces depicting the de novo exchange of c.1766C>T (upper panel) in the proband of pedigree 1 and c.1052T>C (lower panel) in the proband of pedigree 3.
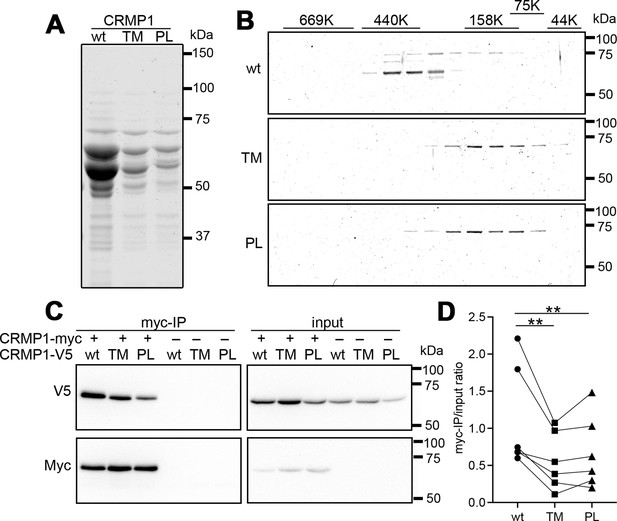
Attenuated oligomer formation of CRMP1-P475L and CRMP1B-T313M variants.
(A) Purified CRMP1B-wildtype, -T313M, and -P475L recombinant proteins on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) stained with CBB. GST-tagged CRMP1 were expressed in E. coli and purified through the binding to glutathione resin and the digestion with PreScission protease. Equal volume (2.5 µl) of the purified specimens, which were prepared under the same condition, were loaded on the gel. The yield of the variants was less than that of CRMP1-wildtype as shown in panel. Two major 64 and 60 kDa bands in the preparations are full-length and a truncated form, respectively. (B) Fractionation of CRMP1B specimens by size-exclusion chromatography. Purified CRMP1B specimens (150 µg) were passed through a Sephacryl S-300 column and the resultant flows/elution volumes were fractionated at every 1 ml. SDS–PAGE for 40–86th fractions were carried out after 20 times concentration. The distribution of molecular weights of standard proteins (44–660 kDa) at the same condition was displayed on the top of the gels. (C) Reduced homophylic interaction of CRMP1B variants. HEK293T cells coexpressing Myc-tagged CRMP1B-wildtype and either one of V5-tagged CRMP1B-wildtype, -T313M, or -P475L were analyzed by co-immunoprecipitation with anti-Myc-antibody. Immunoprecipitated specimens and input lysates were subjected to anti-V5 and anti-Myc immunoblot analyses. Co-immunoprecipitation of V5-tagged CRMP1 variants were reduced comparing to of wildtype CRMP1-V5. (D) Quantification of the V5-signal of Myc-immunoprecipitated specimens and of input lysate. The V5-signal ratios of CRMP1B-T313M and CRMP1B-P475L were significantly decreased compared to the ratio of CRMP1B-wildtype. The graph represents V5-signal ratio of each condition from six independent experiments (n=6). Data were analyzed by one-way repeated measures analysis of variance (ANOVA) followed by Tukey’s multiple comparisons test. **p < 0.01. Abbreviations: CRMP1B-wildtype, wt; CRMP1B-T313M, TM; CRMP1B-P475L, PL.
-
Figure 2—source data 1
CRMP1 variants impact the homo-oligomerization.
- https://cdn.elifesciences.org/articles/80793/elife-80793-fig2-data1-v2.zip
-
Figure 2—source data 2
Ratio of signal intensity of myc-IP band and input ratio of V5 blot.
- https://cdn.elifesciences.org/articles/80793/elife-80793-fig2-data2-v2.zip
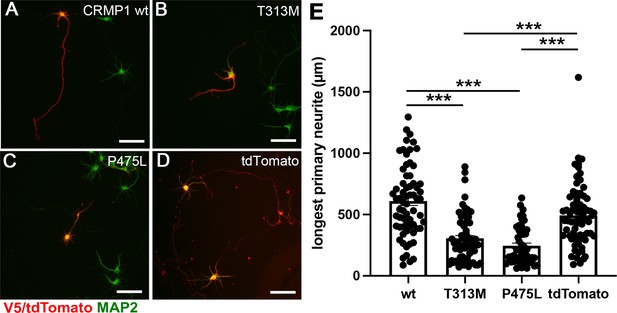
Attenuated neurite outgrowth by the ectopic expression of CRMP1B- P475L and CRMP1B-T313M variants.
Representative images of the neurons expressing V5-CRMP1B-wildtype (A), -T313M (B), -P475L (C), or tdTomato (D). Transfected neurons were visualized by anti-V5 immunostaining or tdTomato expression (red) and anti-MAP2 immunostaining (green). The longest primary neurites of the neurons expressing V5-CRMP1B-T313M or -P475L were shorter than those of the neurons transfected with V5-CRMP1B-wildtype or tdTomato. Scale bars, 100 μm. (E) Longest primary neurite length. The length of the longest primary neurite from V5- or tdTomato-positive neurons was scored in each condition. The graph represents average ± standard error of the mean (SEM) with individual values from four independent experiments. The number (n) of examined neurons in each condition: CRMP1B-wildtype, 66; -T313M, 64; -P475L, 49; tdTomato, 73. Data were analyzed by one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test. ***p < 0.001.
-
Figure 3—source data 1
Quantification of longest neurite length.
- https://cdn.elifesciences.org/articles/80793/elife-80793-fig3-data1-v2.zip
Tables
Phenotype of patients with CRMP1 variants.
| Characteristics and symptoms | Proband 1 (pedigree 1) | Proband 2 (pedigree I1) | Proband 3 (pedigree I1I) |
|---|---|---|---|
| CRMP1 variant (NM_001014809.2); (NP_001014809.1) | c.1766C>T; p.P589L | c.1280C>T; p.T427M | c.1052T>C; p.(F351S) |
| Parents | Non-consanguineous | Non-consanguineous | Non-consanguineous |
| Gender | Female | Male | Female |
| Age at last assessment (years) | 15 | 10 | 13 |
| Anthropometric data | Normal | Normal | Overgrowth |
| Weight (kg) | 57.1 (0.15 SD) | N/A | 133 (14.2 SD) |
| Height (cm) | 168 (0.43 SD) | 166.5 (2.94 SD) | |
| OFC (cm) | 53.5 (−1.02 SD) | 62 (5 SD) | |
| Pregnancy, birth, postnatal adaption | Normal | Normal | Normal |
| singular umbilical artery | + | − | − |
| macrosomia | − | + | − |
| fetal fingerpads | − | + | − |
| Microcephaly (OFC <−2 SD) | − | − | − |
| Macrocephaly (OFC >−2 SD) | − | − | + (5 SD) |
| Facial dysmorphism | − | − | + |
| Delayed motor development | + | + | + |
| walking unsupported | 28 months | 24 months | 24 months |
| Global muscular hypotonia | Mild | Mild | Mild |
| Deep tendon reflexes | Normal | Normal | Normal |
| Intellectual disability | Moderate (IQ 55) | No (IQ 95) | Moderate |
| Autism spectrum disorder | − | + | − |
| Behavioral problems | + | + | + |
| lack of distance, sexualized behavior | + | − | − |
| hyperphagia | − | + | + |
| Delayed speech and language development (first words spoken) | + (24 months) | + (36 months) | + (30 months) |
| Fine motor problems | + | − | + |
| Other | |||
| secondary enuresis | |||
| Nocturna | − | + | + |
| pes planus | − | + | − |
| obesity | − | + | + |
| EEG results | Normal | Normal | Normal |
| Cranial MRI abnormalities | − | − | − |
-
+, yes; −, no; IQ, intellectual quotient; N/A, not available; OFC, occipitofrontal circumference; SD, standard deviation.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Homo sapiens) | CRMP1B | Genbank | NM_001313 transcript variant 2, mRNA | Short isoform of human CRMP1 |
| Strain, strain background (Escherichia coli) | BL21 (DE3) pLysS | BioDynamics | Cat# DS260 Lot 0Q041 | |
| Genetic reagent (M. musculus, female) | ICR | Nihon SLC | RRID:IMSR_TAC:icr | |
| Cell line (Homo sapiens) | HEK293T | ATCC | CRL-3216 | Authenticated by STR analysis |
| Transfected construct (Homo sapiens) | pc3.1beta2-human CRMP1B-V5 wildtype | This paper | pc3.1b2-hCRMP1B-V5 | PCR-amplified from Invitrogen human brain cDNA library Available from F. Nakamura’s lab |
| Transfected construct (Homo sapiens) | pc3.1beta2-human CRMP1B-V5 T313M | This Paper | pc3.1b2-T313M-V5 | Available from F. Nakamura’s lab |
| Transfected construct (Homo sapiens) | pc3.1beta2-human CRMP1B-V5 P475L | This Paper | pc3.1b2-P475L-V5 | Available from F. Nakamura’s lab |
| Transfected construct (Homo sapiens) | pGEX-human CRMP1B wildtype, T313M, P475L | This Paper | pGEX-hCRMP1Bwt, pGEX-T313M, pGEX-P475L | GST-fusion protein expression vectors Available from F. Nakamura’s lab |
| Transfected construct (Discosoma sp.) | pENN.AAV.CAG.tdTomato.WPRE.SV40 | Gift from James M. Wilson (Addgene plasmid) | # 105554; RRID:Addgene_105554 | — |
| Antibody | Anti-MAP2 (Rabbit polyclonal) | Covance | PRB−547C RRID:AB_2565455 | 1:5000 |
| Antibody | Anti-Myc-tag (9E10, mouse monoclonal) conjugated agarose resin | BD | Cat# 631208 Lot 4100006 | 1:1000 |
| Antibody | Anti-Myc-tag (My3) (Mouse monoclonal) | MBL | Cat# M192-3 Lot 009 | 1:5000 |
| Antibody | Anti-V5 tag (Mouse monoclonal) | ThermoFisher | Cat# R960-25 RRID:AB_2556564 Lot 2311401 | 1:5000 |
| Antibody | Anti-V5 tag (Rabbit polyclonal) | Novus Biological | Cat# NB600-381 RRID:AB_527427 | 1:5000 |
| Antibody | Anti-mouse Immunogloblins/HRP (goat polyclonal) | GE | Cat# NA931V | 1:5000 |
| Antibody | Anti-mouse Immunogloblins/ biotin (goat polyclonal) | Jackson | Cat# P0260 | 1:5000 |
| Antibody | Anti-rabbit Immunogloblins/HRP (goat polyclonal) | GE | Cat# NA934V | 1:5000 |
| Antibody | Anti-rabbit Immunogloblins/biotin (goat polyclonal) | Vector | Cat# P0260 | 1:5000 |
| Recombinant DNA reagent | pc3.1beta2-V5 | Kawashima et al., 2021 (J.Neurochem 157: 1207–1221 (2021)) | N/A | |
| Recombinant DNA reagent | pGEX-6P-1 | Cytiva | Cat# 28954648 | |
| Sequence-based reagent | Human CRMP1B-1f primer | This paper | PCR primers | atcgaattcgccATGTCGTACCAGGGCAAGAAGAGCAT |
| Sequence-based reagent | Human CRMP1-572r primer | This paper | PCR primers | atcctcgagACCGAGGCTGGTGATGTTGGAGCGGCCACCA |
| Sequence-based reagent | T313M mutation primer | This paper | Mutation primers | CCGGACCCTACCAtGCCCGACTACTTG |
| Sequence-based reagent | P475L mutation primer | This paper | Mutation primers | CGGAAGGCGTTCCtGGAGCACCTGTAC |
| Sequence-based reagent | CRMP1-forward primer (P1) | This paper | Sanger sequencing primers | TCTTCGAGGGTATGGAGTGC |
| Sequence-based reagent | CRMP1-reverse primer (P1) | This paper | Sanger sequencing primers | CGTCAGATCTCGATTCCCCA |
| Sequence-based reagent | CRMP1-forward primer (P2) | This paper | Sanger sequencing primers | ACAAAAGCGGATCCTGGAGA |
| Sequence-based reagent | CRMP1-reverse primer (P2) | This paper | Sanger sequencing primers | GTACACAGGGCAGTTGATCC |
| Peptide, recombinant protein | PreScission protease | Cytiva | Cat# 27084301 | |
| Peptide, recombinant protein | hCRMP1B-wt, T313M, P475L | This paper | Purified from E. coli BL21 (DE3) pLysS cells Available from F. Nakamura’s lab | |
| Commercial assay or kit | QuickChange Multi Site-Directed Mutagenesis Kit | Agilent Tech | Cat #200515-5 | |
| Commercial assay or kit | Tyramide signal amplification (TSA) system | PerkinElmer | Cat No. NEL700A001KT | |
| Chemical compound, drug | Glutathione Sepharose 4B, 10 ml | Cytiva | Cat# 17075601 | |
| Chemical compound, drug | Poly-L-lysine | Wako | Cat No. 163-19091 | |
| Chemical compound, drug | Fugene-6 | Promega | Cat No. E2691 | |
| Software, algorithm | Fiji (2.0.0-rc-59/1.51n) | Schindelin et al., 2012 Fiji software (2.0.0-rc-59/1.51n) | https://imagej.net/software/fiji/ | |
| Software, algorithm | Prism (Version 9.4.1) | GraphPad Software, LLC. | https://www.graphpad.com/scientific-software/prism/ |
Additional files
-
Supplementary file 1
Short tandem repeat (STR) profile and mycoplasma contamination test for HEK293T cell line.
- https://cdn.elifesciences.org/articles/80793/elife-80793-supp1-v2.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/80793/elife-80793-mdarchecklist1-v2.pdf





