Endosymbiotic selective pressure at the origin of eukaryotic cell biology
Figures
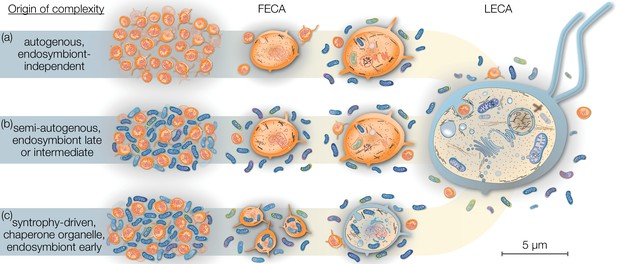
Eukaryogenesis scenarios with respect to the origin and nature of the first eukaryotic common ancestor (FECA).
Scenarios summarized in (a) were based on the assumption that eukaryotic groups existed that lack mitochondria and which originated independently of endosymbiosis. Such models are no longer supported and we understand that the last eukaryotic common ancestor (LECA) was a mitochondrion-bearing lineage. Models in (b) speculate on a (semi-) autogenous origin of complexity. By their definition, the FECA was free of vertically transmittable endosymbionts. Genomic and cellular complexity of FECA grew through horizontally donated genes from diverse prokaryotes part of its habitat; to a degree FECA was eukaryote-like prior to endosymbiosis. Models falling under (c) differ from (b) in that both partners at the onset of eukaryogenesis were prokaryotic in nature and that complexity only began to evolve after endosymbiosis of the mitochondrial ancestor. By this definition, the FECA is characterized by the endosymbiotic event and describes the lineage that transitioned from prokaryotic to eukaryotic evolution. Note that the prokaryotic cells on the left and the LECA on the right are shown to scale on the basis of average cell diameters.
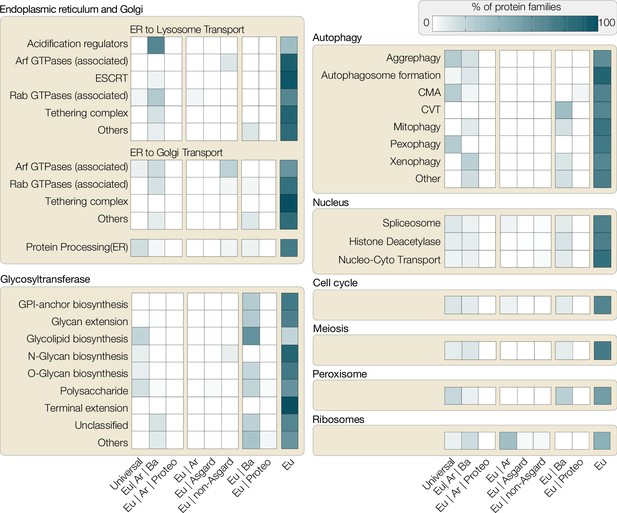
Distribution of protein families involved in eukaryotic traits.
All eukaryotic protein sequences associated with different functions in KEGG (Kyoto Encyclopedia of Genes and Genomes, as of November 2021) were used to build representative hidden Markov models (HMMER/3.1), which were then searched against all prokaryotic genomes available on KEGG. Based on hits found across bacterial phyla, the following categories were assigned: Universal (shared with >50% bacterial phyla and archaea), Eu|Ar|Ba (shared with archaea and <50% bacterial phyla), Eu|Ar|Pr (shared with archaea and proteobacteria only), Eu|Ar (shared with Ar), Eu|Asgard (shared only with asgard archaea), Eu|non-Asgard (shared only with non-asgard archaea), Eu|Ba (shared with bacteria), Eu|Proteo (shared only with proteobacteria), Eu (not shared with any prokaryotes). Each box shows the percentage of protein families across these categories (x-axis) for the pathways analysed. For example, a substantial percentage of ribosomal proteins (bottom right) are shared between Eukaryote-Archaea or Eukaryote-Archaea-Bacteria, highlighting the host’s ribosomal contribution to eukaryotes. Proteins for other categories, however, are either predominantly eukaryote-specific or shared between eukaryotes and bacteria (e.g. glycotransferases or some proteins of autophagy). CMA, chaperone-mediated autophagy; CVT, cytoplasm to vacuolar targeting pathway.
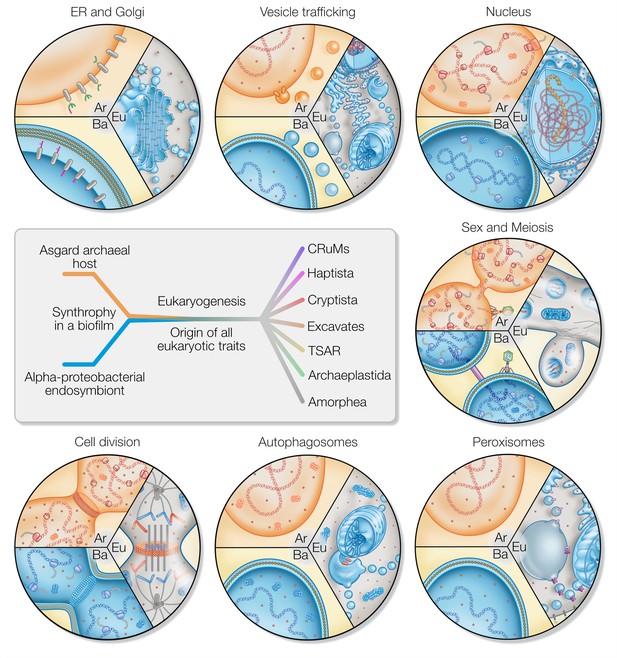
Eukaryotic traits in light of housing an endosymbiont.
Each segment highlights an eukaryotic (Eu) trait and the comparable, if present, situation in bacteria (Ba) and archaea (Ar). (i) Prokaryotes secrete outer membrane vesicles (OMVs) and an endosymbiont (mitochondrion) secreting OMVs could have (ii) given rise to a dynamic endomembrane system within the archaeal host and explaining the transition from archaeal to bacterial lipids. N-glycosylation has been identified in all domains of life, but the eukaryotic N-glycosylation pathway is homologous to that of archaea. (iii) A specialized extension of the endoplasmic reticulum (ER), the nucleus prevents co-transcriptional translation of proteins – as is the rule in prokaryotes – to allow for the splicing of introns. (iv) Prokaryotes constantly shed and acquire DNA from the environment, and often promiscuously by transformation, transduction, and conjugation. In the absence of such dedicated mechanisms, eukaryotes avoid Muller’s ratchet through sex and meiosis whose origin might be linked to coordinating the merging of two genomes and synchronizing nuclear and mitochondrial division. (v) Peroxisomes also form through mitochondria-derived vesicles and house enzymes of alphaproteobacterial origin. (vi) Eukaryotes perform autophagy using membranes and proteins of the ESCRT machinery to surround and digest internal membrane compartments including the mitochondrion. (vii) While bacteria use homologs of tubulin to perform fission, eukaryotic fission utilizes actin and components of the ESCRT machinery similar to archaea, whereas the tubulin in eukaryotes is for instance used to separate chromatin and intracellular compartments.
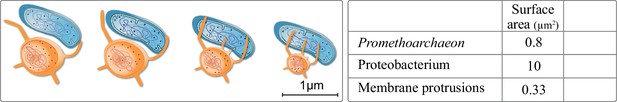
Extracellular membrane protrusions from asgard archaea.
Potential trade-off between cytosol investment between cell and protrusion. Each 1 µm increase in protrusion takes 12% cytoplasm, likely limiting total protrusion and reducing cell volume. Values in the table for the asgard archaea were calculated based on Eme et al., 2017: average asgard cell radius: 0.25 µm, average protrusion radius 50 nm, and length 1 µm. Average alphaproteobacterium radius and length were taken to be 0.5 and 3 µm, respectively.
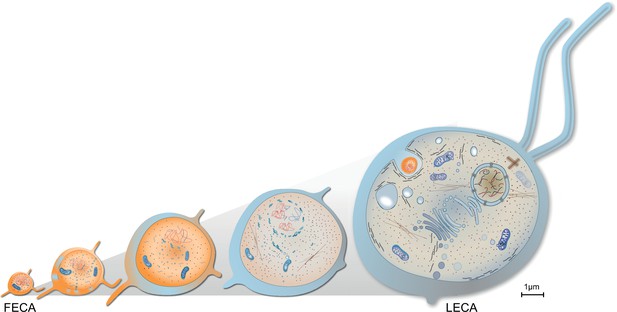
Evolution of last eukaryotic common ancestor (LECA) after the symbiotic event.
Vesicles from the proteobacterium accumulated in cytosol giving rise to permeative barrier around the host DNA Brueckner and Martin, 2020, selected positively due to parallel bombardment of genes (Land et al., 2015). As a result, the genome of bacterium shrunk, whereas that of the host expanded, along with increase in cell size. New combinations of genes, powered by energy from the endosymbiont, led to emergence of novel trait and LECA with eukaryotic cell biology.
Tables
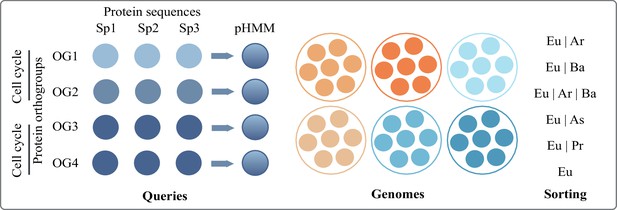
Key to the different groups compared among each other.
| Category | Present in |
|---|---|
| Universal | >50% bacterial phyla, asgard and non-asgard archaea |
| Eu|Ar|Ba | <50% bacterial phyla, asgard and non-asgard archaea |
| Eu|Ar|Proteo | Asgard and non-asgard archaea, exclusively proteobacteria (no other bacterial phylum) |
| Eu|Ar | Asgard and non-asgard archaea, no bacterial phylum |
| Eu|Asgard | Asgard only (no other archaea or bacteria) |
| Eu|non-Asgard | Non-asgard only (no other bacteria or asgard archaea) |
| Eu|Ba | Bacterial phylum other than proteobacteria |
| Eu|Proteo | Only proteobacterial phyla |
| Eukaryotes (Eu) | No hits in >5% of any prokaryotic phylum |