A parasitic fungus employs mutated eIF4A to survive on rocaglate-synthesizing Aglaia plants
Figures
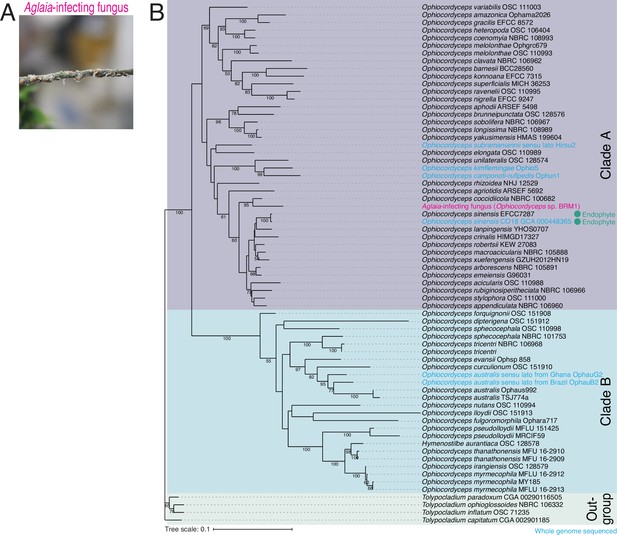
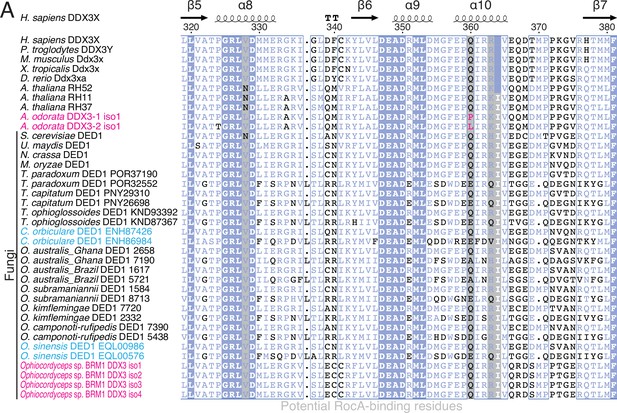
Identification of Aglaia-parasitic Ophiocordyceps sp. BRM1.
(A) Image of a parasite fungus growing on Aglaia odorata. (B) Multilocus phylogenetic tree of Ophiocordyceps species generated from maximum likelihood phylogenetic analysis of ITS, SSU, LSU, RPB1, and TEF1α sequences. Tolypocladium species were used as outgroups. The best DNA substitution models of ITS, LSU, SSU, RPB1, and TEF1α were calculated as TIM3ef + G4, TIM1 + I + G4, TIM3ef + I + G4, TrN + I + G4, and TIM1 + I + G4, respectively. Numbers on branches are percent support values out of 1000 bootstrap replicates. Only bootstrap values greater than 50% support are shown. Endophytes are highlighted with green dots.
-
Figure 1—source data 1
Files for the full and unedited pictures corresponding to Figure 1A.
- https://cdn.elifesciences.org/articles/81302/elife-81302-fig1-data1-v1.zip
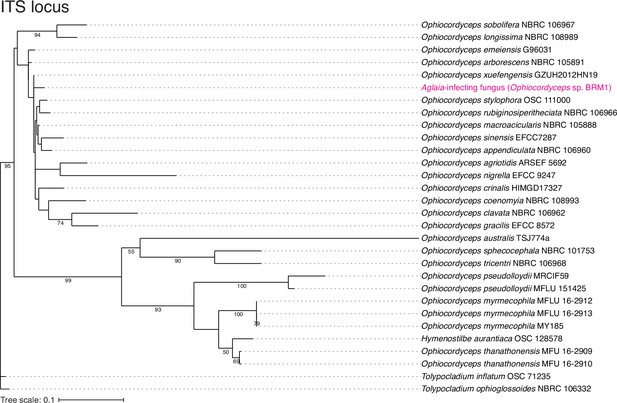
Assessment of Aglaia-infecting fungus species by ITS locus.
The maximum likelihood best scoring trees based on ITS locus from the Ophiocordyceps species with Tolypocladium species as outgroups. Numbers at nodes are percentages of bootstrap support values out of 1000. Only bootstrap values above 50% are shown.
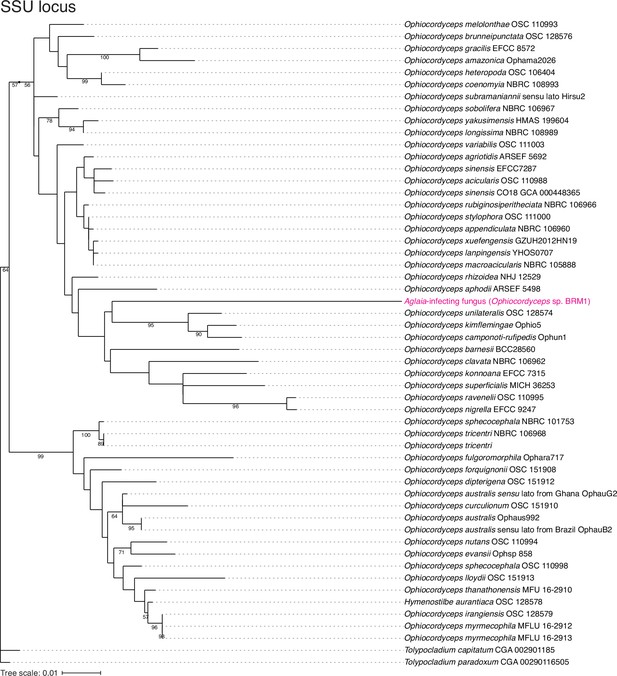
Assessment of Aglaia-infecting fungus species by SSU locus.
The maximum likelihood best scoring trees based on SSU locus from the Ophiocordyceps species with Tolypocladium species as outgroups. Numbers at nodes are percentages of bootstrap support values out of 1,000. Only bootstrap values above 50% are shown.
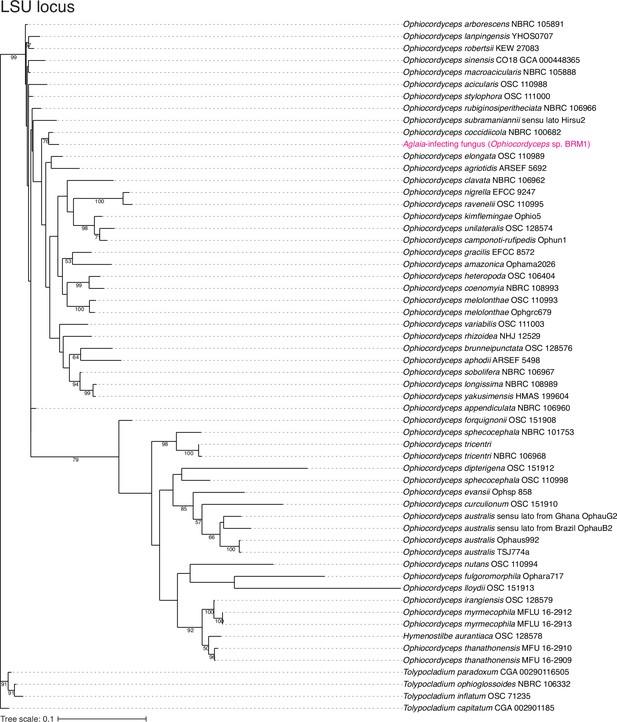
Assessment of Aglaia-infecting fungus species by LSU locus.
The maximum likelihood best scoring trees based on LSU locus from the Ophiocordyceps species with Tolypocladium species as outgroups. Numbers at nodes are percentages of bootstrap support values out of 1000. Only bootstrap values above 50% are shown.
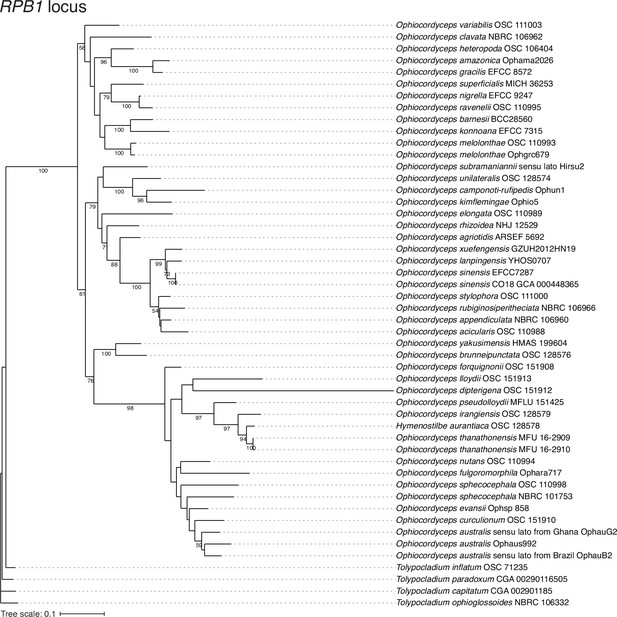
Assessment of Aglaia-infecting fungus species by RPB1 locus.
The maximum likelihood best scoring trees based on RPB1 locus from the Ophiocordyceps species with Tolypocladium species as outgroups. Numbers at nodes are percentages of bootstrap support values out of 1000. Only bootstrap values above 50% are shown.
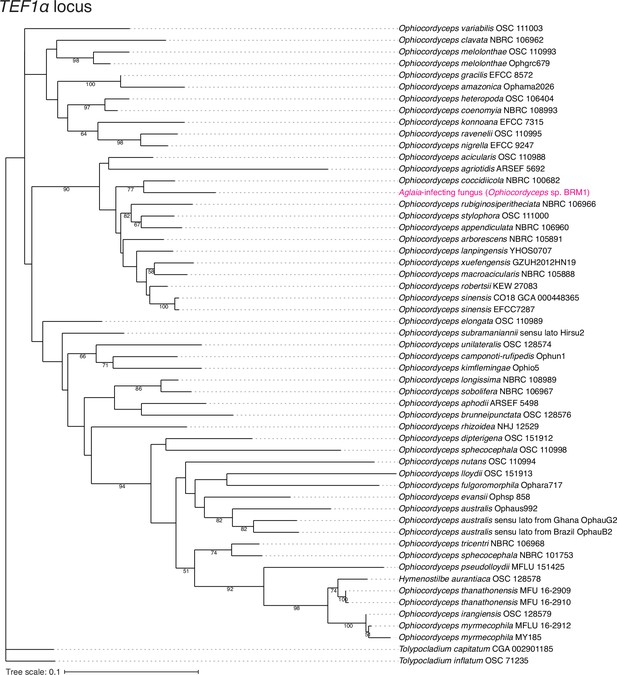
Assessment of Aglaia-infecting fungus species by TEF1α locus.
The maximum likelihood best scoring trees based on TEF1α locus from the Ophiocordyceps species with Tolypocladium species as outgroups. Numbers at nodes are percentages of bootstrap support values out of 1000. Only bootstrap values above 50% are shown.
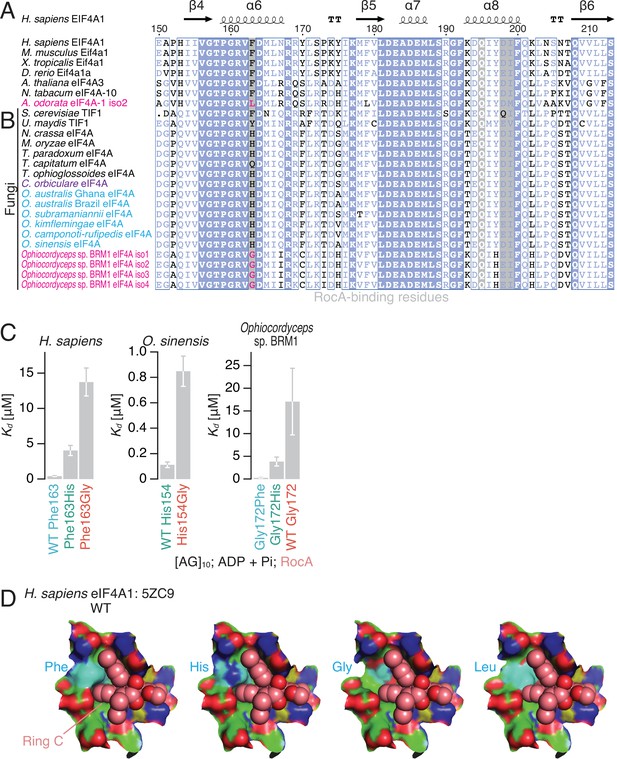
The effect of an amino acid substitution found in Ophiocordyceps sp. BRM1 eIF4A on RocA-mediated polypurine RNA clamping.
(A, B) Alignments of eIF4A protein sequences from higher eukaryotes (A) and fungal species (B), including the de novo-assembled Ophiocordyceps sp. BRM1 eIF4A gene with four transcript isoforms (iso). (C) The summary of Kd determined by fluorescence polarization assay in Figure 2—figure supplement 2A–H is depicted. WT and mutated eIF4A proteins from the indicated species were used. To measure ATP-independent RNA clamping induced by RocA (50 µM), ADP and Pi (1 mM each) were included in the reaction. The data are presented as the mean and s.d. values. (D) RocA (sphere model with light pink-colored carbons), the modeled His, Gly, and Leu residues (surface model with cyan-colored carbons) at the Phe163 residue in human eIF4A1 (surface model with green-colored carbons), and RNA (surface model with yellow-colored carbons) in the complex of human eIF4A1•RocA•AMP-PNP•polypurine RNA (PDB: 5ZC9) (Iwasaki et al., 2019).
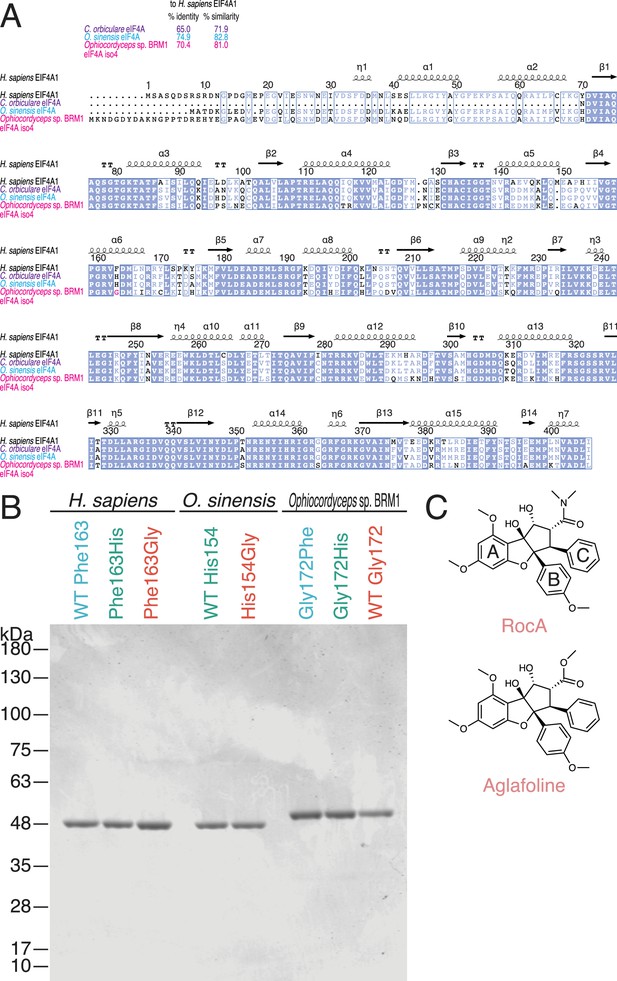
Characterization of recombinant proteins used in this study.
(A) Alignment of eIF4A protein sequences from the indicated species. Percent similarity and percent identity to H. sapiens eIF4A1 are shown at the top. (B) Coomassie brilliant blue staining of recombinant eIF4A proteins used in this study. (C) Chemical structures of rocaglates used in this study.
-
Figure 2—figure supplement 1—source data 1
Files for the full and unedited gel images corresponding to Figure 2—figure supplement 1B.
- https://cdn.elifesciences.org/articles/81302/elife-81302-fig2-figsupp1-data1-v1.zip
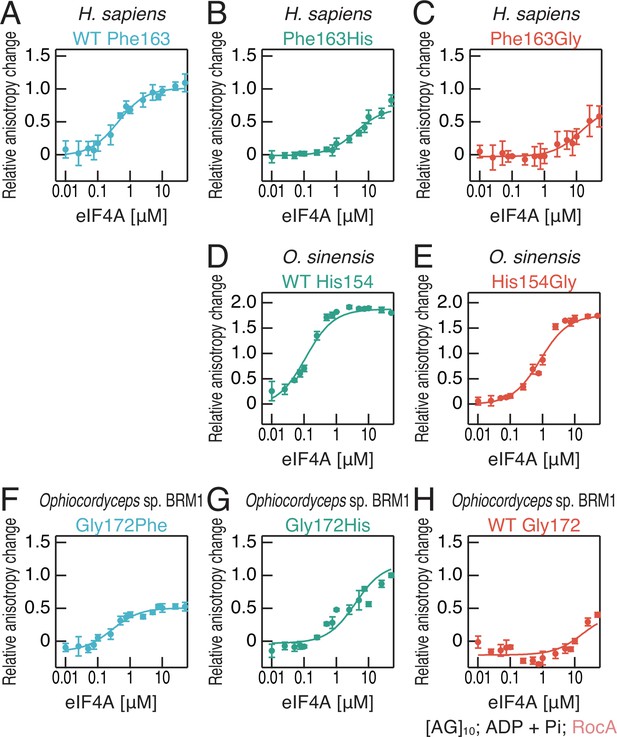
Affinities between polypurine RNA and recombinant eIF4A proteins in the presence of RocA, ADP, and Pi.
(A–H) Fluorescence polarization assay for FAM-labeled RNA ([AG]10) (10 nM). WT and mutated eIF4A proteins from the indicated species were used. To measure ATP-independent RNA clamping induced by RocA (50 µM), ADP and Pi (1 mM each) were included in the reaction. The data are presented as the mean and s.d. values (n = 3).
-
Figure 2—figure supplement 2—source data 1
Files for the primary data corresponding to Figure 2—figure supplement 2A–H.
- https://cdn.elifesciences.org/articles/81302/elife-81302-fig2-figsupp2-data1-v1.zip
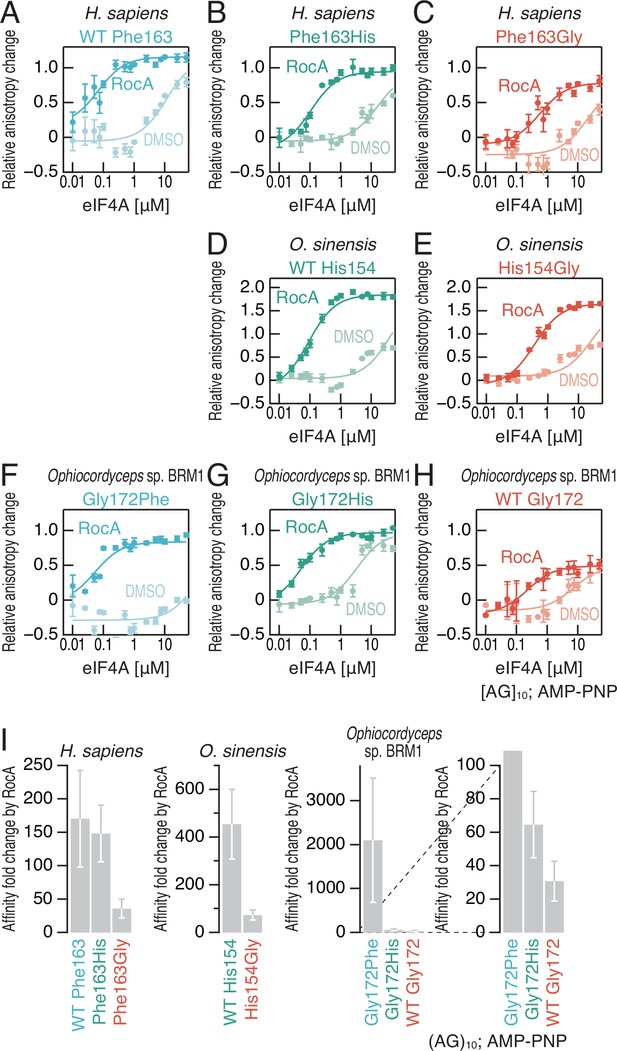
Affinities between polypurine RNA and recombinant eIF4A proteins in the presence of an ground-state ATP analog.
(A–H) Fluorescence polarization assay for FAM-labeled RNA ([AG]10) (10 nM). WT and mutated eIF4A proteins from the indicated species were used. The ground-state ATP analog AMP-PNP (1 mM) was included in the reaction with RocA (50 µM). The data are presented as the mean and s.d. values (n = 3). (I) Affinity fold changes between DMSO and RocA treatment by amino acid substitutions in (A–H) were calculated. The data are presented as the mean and s.d. values.
-
Figure 2—figure supplement 3—source data 1
Files for the primary data corresponding to Figure 2—figure supplement 3A–H.
- https://cdn.elifesciences.org/articles/81302/elife-81302-fig2-figsupp3-data1-v1.zip
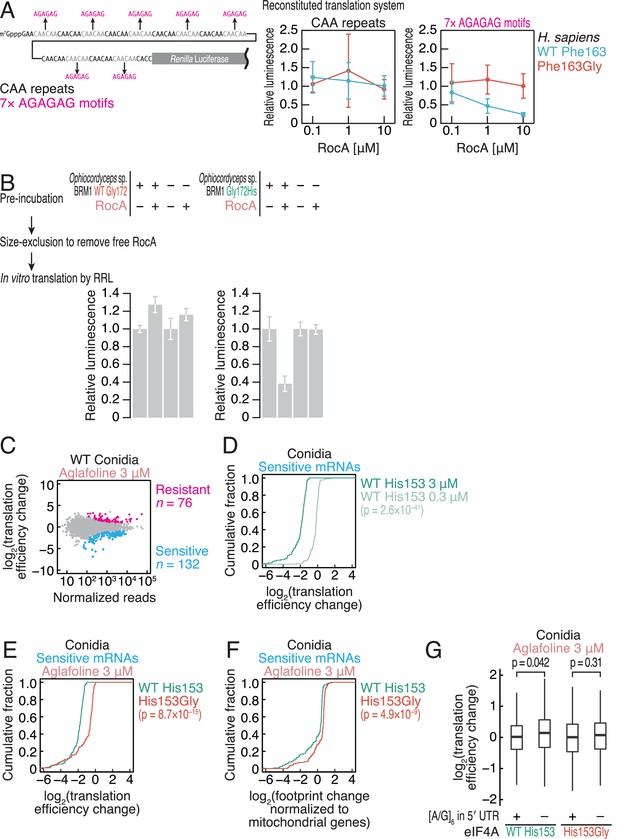
The amino acid substitution in the Ophiocordyceps sp. BRM1 eIF4A confers translational resistance to rocaglates in fungi.
(A) RocA-mediated translational repression recapitulated by an in vitro reconstitution system with human factors. Recombinant proteins of H. sapiens eIF4A1 WT or Phe163Gly were added to the reaction with RocA. Reporter mRNA with CAA repeats or polypurine motifs was translated in the reaction. The data are presented as the mean and s.d. values (n = 3). (B) Translation of complex-preformed mRNAs to test the RocA gain of function. Recombinant proteins of Ophiocordyceps sp. BRM1 eIF4A1 WT or the Gly172His mutant were preincubated with the reporter mRNA possessing polypurine motifs in the presence or absence of RocA. After removal of free RocA by gel filtration, the protein-mRNA complex was added to RRL to monitor protein synthesis. The data are presented as the mean and s.d. values (n = 3). (C) MA (M, log ratio; A, mean average) plot of the translation efficiency changes caused by 3 µM aglafoline treatment in C. orbiculare eIF4AWT conidia. Resistant and sensitive mRNAs (FDR < 0.05) are highlighted. (D) Cumulative distribution of the translation efficiency changes in aglafoline-sensitive mRNAs (defined in C) in C. orbiculare eIF4AWT conidia treated with 0.3 or 3 µM aglafoline. (E) Cumulative distribution of the translation efficiency changes in aglafoline-sensitive mRNAs (defined in C) induced by 3 µM aglafoline treatment in C. orbiculare eIF4AWT and eIF4AHis153Gly conidia. (F) Cumulative distribution of the global translation alterations, which are footprint changes normalized to mitochondrial footprints, in aglafoline-sensitive mRNAs (defined in C) induced by 3 µM aglafoline treatment in C. orbiculare eIF4AWT and eIF4AHis153Gly conidia. (G) Box plot of the translation efficiency changes caused by 3 µM aglafoline treatment in conidia across mRNAs with or without an [A/G]6 motif in the 5′ UTR. The p values in (D–G) were calculated by the Mann–Whitney U test.
-
Figure 3—source data 1
Files for the primary data corresponding to Figure 3A.
- https://cdn.elifesciences.org/articles/81302/elife-81302-fig3-data1-v1.zip
-
Figure 3—source data 2
Files for the primary data corresponding to Figure 3B.
- https://cdn.elifesciences.org/articles/81302/elife-81302-fig3-data2-v1.zip
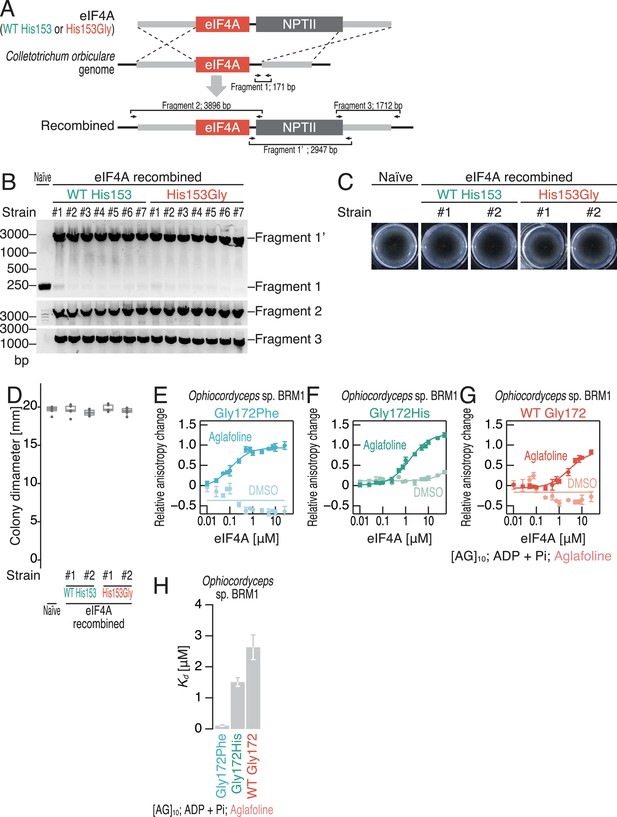
Establishment of eIF4A-engineered C. orbiculare strains.
(A) Schematics of eIF4A recombination in C. orbiculare. NPTII, neomycin phosphotransferase II. (B) PCR-based screening of the recombined strains. The primer sets used for screening are depicted in (A). (C, D) Colony formation of the indicated C. orbiculare strains cultured in PDA for 5 days (C). The measured colony diameters are shown in the box plot (n = 5) (D). (E–G) Fluorescence polarization assay for FAM-labeled RNA ([AG]10) (10 nM). WT and mutated Ophiocordyceps sp. BRM1 eIF4A proteins were used. To measure ATP-independent RNA clamping induced by aglafoline (50 µM), ADP and Pi were included in the reaction. The data are presented as the mean and s.d. values (n = 3). (H) Summary of the Kd values in (E–G) under treatment with aglafoline. The data are presented as the mean and s.d. values.
-
Figure 3—figure supplement 1—source data 1
Files for the full and unedited gel images corresponding to Figure 3—figure supplement 1B and C.
- https://cdn.elifesciences.org/articles/81302/elife-81302-fig3-figsupp1-data1-v1.zip
-
Figure 3—figure supplement 1—source data 2
Files for the primary data corresponding to Figure 3—figure supplement 1D.
- https://cdn.elifesciences.org/articles/81302/elife-81302-fig3-figsupp1-data2-v1.zip
-
Figure 3—figure supplement 1—source data 3
Files for the primary data corresponding to Figure 3—figure supplement 1E–G.
- https://cdn.elifesciences.org/articles/81302/elife-81302-fig3-figsupp1-data3-v1.zip
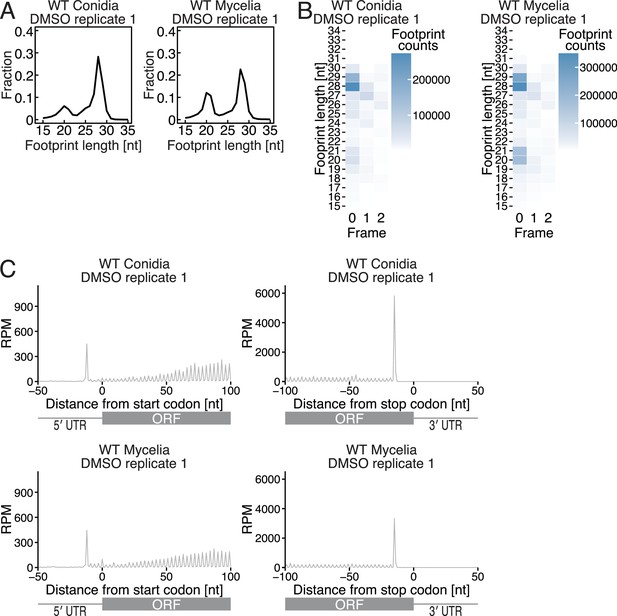
Characterization of ribosome footprints in C. orbiculare.
(A) Distribution of ribosome footprint length in conidia and mycelia. (B) Tile plot of reading frames at each ribosome footprint length in conidia and mycelia. The 5′ end positions of the ribosome footprints are depicted. The footprint count scales are shown in the color bars. (C) Metagene plot of 29-nt ribosome footprints around start (left) and stop (right) codons in conidia and mycelia. The 5′ end positions of the ribosome footprints are depicted. RPM: reads per million mapped reads.
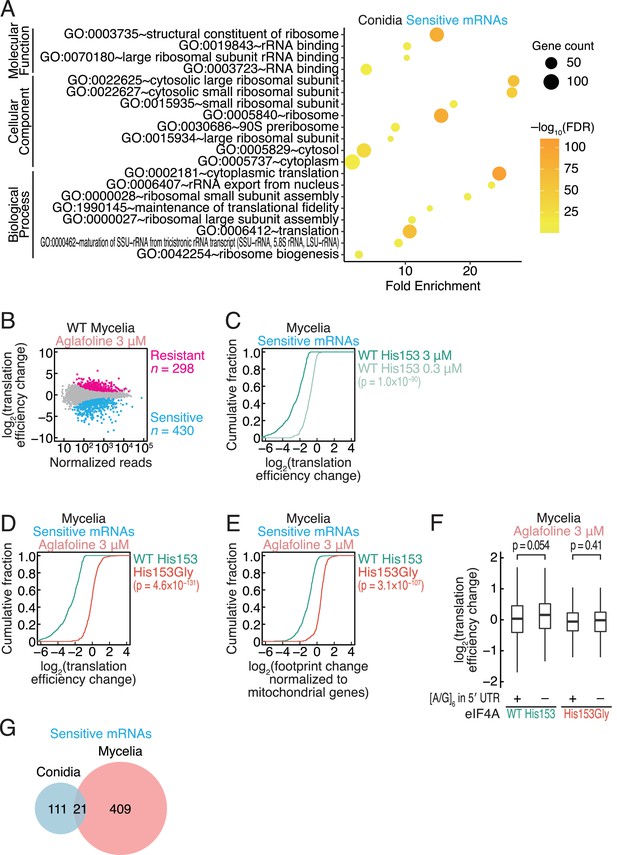
Translation changes by aglafoline treatment in recombined C. orbiculare.
(A) GO term analysis of aglafoline-sensitive mRNAs (defined in Figure 3C). GO terms associated with yeast homologs were analyzed by DAVID (Huang et al., 2009a; Huang et al., 2009b). (B) MA plot of the translation efficiency changes induced by 3 µM aglafoline treatment in C. orbiculare eIF4AWT mycelia. Resistant and sensitive mRNAs (false discovery rate [FDR] < 0.05) are highlighted. (C) Cumulative distribution of the translation efficiency changes in aglafoline-sensitive mRNAs (defined in B) in C. orbiculare eIF4AWT mycelia treated with 0.3 or 3 µM aglafoline. (D) Cumulative distribution of the translation efficiency changes in aglafoline-sensitive mRNAs (defined in B) induced by 3 µM aglafoline treatment in C. orbiculare eIF4AWT and eIF4AHis153Gly mycelia. (E) Cumulative distribution of the global translation alterations, which are footprint changes normalized to mitochondrial footprints, in aglafoline-sensitive mRNAs (defined in B) induced by 3 µM aglafoline treatment in C. orbiculare eIF4AWT and eIF4AHis153Gly conidia. (F) Box plot of translation efficiency changes caused by 3 µM aglafoline treatment in mycelia across mRNAs with or without an [A/G]6 motif in the 5′ UTR. (G) Venn diagram of the overlap between aglafoline-sensitive mRNAs in conidia (defined in Figure 3C) and mycelia (defined in B). The p values in (C–F) were calculated by the Mann–Whitney U test.
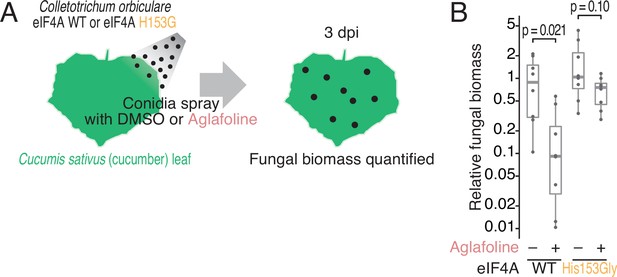
Phenotypic comparison of the C. orbiculare eIF4AWT and eIF4AHis153Gly strains during infection in the presence of rocaglate.
(A) Workflow for monitoring the biomass of C. orbiculare eIF4AWT or eIF4AHis153Gly strains on cucumber leaves under treatment with aglafoline. (B) Comparison of in planta fungal biomass of C. orbiculare eIF4AWT or eIF4AHis153Gly strains with or without treatment with 1 µM aglafoline. Relative expression levels of the C. orbiculare 60 S ribosomal protein L5 gene (GenBank: Cob_v012718) normalized to that of a cucumber cyclophilin gene (GenBank: AY942800.1) were determined by RT–qPCR at 3 dpi (n = 8). The relative fungal biomasses of C. orbiculare were normalized to those of eIF4AWT without aglafoline. Significance was calculated by Student’s t-test (two-tailed). Three independent experiments showed similar results.
-
Figure 4—source data 1
Files for the primary data corresponding to Figure 4B.
- https://cdn.elifesciences.org/articles/81302/elife-81302-fig4-data1-v1.zip

Characterization of cucumber leaves treated with aglafoline.
(A) C. sativus leaves were sprayed with DMSO or aglafoline (1 µM) in water and incubated for 3 days using the same method as C. orbiculare inoculation.
-
Figure 4—figure supplement 1—source data 1
Files for the full and unedited pictures corresponding to Figure 4—figure supplement 1A.
- https://cdn.elifesciences.org/articles/81302/elife-81302-fig4-figsupp1-data1-v1.zip
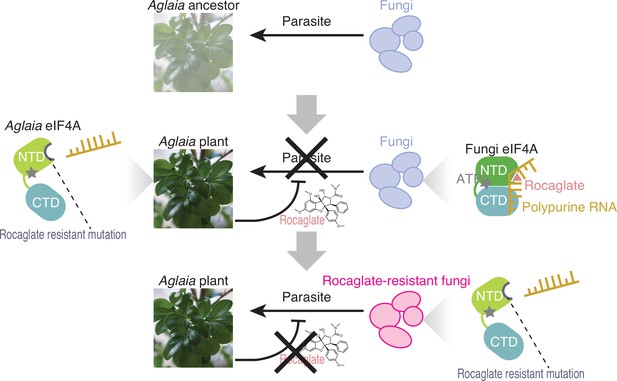
Model of the plant–fungus arms race evoked by rocaglates.
The ancestors of the Aglaia plants may have been subjected to fungal infection. To counteract this, Aglaia plants may have developed rocaglates to target the conserved translation factor eIF4A and to suppress in planta fungal growth. Simultaneously, Aglaia plants exhibit amino acid substitutions in the rocaglate binding pocket of eIF4As to prevent self-poisoning. Some fungi may impede rocaglate toxin by converting eIF4A to a rocaglate-insensitive form, enabling them to parasitize these plants.
Tables
Summary of Kd (µM) between eIF4A protein and RNAs.
A fluorescence polarization assay between FAM-labeled RNA ([AG]10) and the indicated recombinant proteins was conducted to measure Kd in the presence of DMSO, RocA, or aglafoline. ND, not determined.
| [AG]10 | |||||
|---|---|---|---|---|---|
| ADP + Pi | AMP-PNP | ||||
| Protein | DMSO | RocA | Aglafoline | DMSO | RocA |
| H. sapiens WT Phe163 | 0.42 ± 0.061 | 11 ± 2.9 | 0.067 ± 0.023 | ||
| H. sapiens Phe163His | 4.0 ± 0.71 | 16 ± 2.7 | 0.11 ± 0.025 | ||
| H. sapiens Phe163Gly | 14 ± 2.0 | 21 ± 6.7 | 0.58 ± 0.13 | ||
| O.sinensis WT His154 | 0.11 ± 0.022 | 41 ± 11 | 0.090 ± 0.014 | ||
| O.sinensis His154Gly | 0.85 ± 0.12 | 27 ± 7.0 | 0.37 ± 0.046 | ||
| Ophiocordyceps sp. BRM1 Gly172Phe | ND | 0.27 ± 0.050 | 0.11 ± 0.021 | 110 ± 58 | 0.053 ± 0.023 |
| Ophiocordyceps sp. BRM1 Gly172His | ND | 3.9 ± 0.98 | 1.5 ± 0.14 | 3.3 ± 0.83 | 0.051 ± 0.0091 |
| Ophiocordyceps sp. BRM1 WT Gly172 | ND | 17±7.4 | 2.6±0.40 | 7.1±2.3 | 0.23±0.050 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Homo sapiens) | eIF4A1 | NCBI | GenBank:CCDS11113.1 | |
| Gene (Ophiocordyceps sinensis) | eIF4A | EnsemblFungi (https://fungi.ensembl.org/index.html) | EnsemblFungi:OCS_04979 | |
| Gene (Ophiocordyceps sp. BRM1) | eIF4A iso4 | This study | Iwasaki lab | |
| Gene (Colletotrichum orbiculare) | eIF4A | GenBank | GenBank:Cob_v000942 | |
| Gene (C. orbiculare) | 60S ribosomal protein L5 | GenBank | GenBank:Cob_v012718 | |
| Gene (Cucumis sativus) | Cyclophilin | GenBank | GenBank:AY942800.1 | |
| Strain, strain background (Aglaia odorata) | Aglaia odorata | This paper | Grown in Berkeley, CA; Ingolia lab | |
| Strain, strain background (Ophiocordyceps sp. BRM1) | Ophiocordyceps sp. BRM1 | This paper | Grown in Berkeley, CA; Ingolia lab | |
| Strain, strain background (Escherichia coli) | BL21 Star (DE3) | Thermo Fisher Scientific | Cat. #:C601003 | |
| Strain, strain background (C. orbiculare) | 104-T | NARO GenBank | MAFF 240422 | |
| Strain, strain background (C. orbiculare) | eIF4AWT#1 | This paper | CoNK1171 | Supplementary file 4; Shirasu lab |
| Strain, strain background (C. orbiculare) | eIF4AWT#2 | This paper | CoNK1172 | Supplementary file 4; Shirasu lab |
| Strain, strain background (C. orbiculare) | eIF4AH153G#1 | This paper | CoNK1181 | Supplementary file 4; Shirasu lab |
| Strain, strain background (C. orbiculare) | eIF4AH153G#2 | This paper | CoNK1182 | Supplementary file 4; Shirasu lab |
| Strain, strain background (C. sativus) | Suyo strain | Sakata Seed Corp. | ||
| Recombinant DNA reagent | pColdI (plasmid) | TaKaRa | Cat. #:3361 | |
| Recombinant DNA reagent | pColdI-H. sapiens eIF4A1 WT (plasmid) | RIEN BRC | RDB17299 | Iwasaki et al., 2019 |
| Recombinant DNA reagent | pColdI-H. sapiens eIF4A1 Phe163Gly (plasmid) | This paper | Iwasaki lab | |
| Recombinant DNA reagent | pColdI-H. sapiens eIF4A1 Phe163His (plasmid) | This paper | Iwasaki lab | |
| Recombinant DNA reagent | pColdI-O. sinensis eIF4A WT (plasmid) | This paper | Iwasaki lab | |
| Recombinant DNA reagent | pColdI-O. sinensis eIF4A His154Gly (plasmid) | This paper | Iwasaki lab | |
| Recombinant DNA reagent | pColdI-Ophiocordyceps sp. BRM1 eIF4A iso4 WT (plasmid) | This paper | Iwasaki lab | |
| Recombinant DNA reagent | pColdI-Ophiocordyceps sp. BRM1 eIF4A iso4 Gly172His (plasmid) | This paper | Iwasaki lab | |
| Recombinant DNA reagent | pColdI-Ophiocordyceps sp. BRM1 eIF4A iso4 Gly172Phe (plasmid) | This paper | Iwasaki lab | |
| Recombinant DNA reagent | pENTR4 (plasmid) | Thermo Fisher Scientific | Cat. #:A10465 | |
| Recombinant DNA reagent | pENTR4-C. orbiculare eIF4A WT (plasmid) | This paper | Shirasu lab | |
| Recombinant DNA reagent | pENTR4-C. orbiculare eIF4A His153Gly (plasmid) | This paper | Shirasu lab | |
| Recombinant DNA reagent | pII99 (plasmid) | Namiki et al., 2001 | ||
| Recombinant DNA reagent | psiCHECK2−7×AGAGAG motifs | Iwasaki et al., 2016 | ||
| Recombinant DNA reagent | psiCHECK2-CAA repeats | Iwasaki et al., 2016 | ||
| Sequence-based reagent | Random Primer (nonadeoxyribonucleotide mix: pd(N)9) | TaKaRa | Cat. #:3802 | |
| Sequence-based reagent | FAM-labeled [AG]10 RNA | Iwasaki et al., 2019 | ||
| Sequence-based reagent | Primers | This paper | Supplementary file 5; Shirasu lab | |
| Peptide, recombinant protein | H. sapiens eIF4A1 WT | This paper | Iwasaki lab | |
| Peptide, recombinant protein | H. sapiens eIF4A1 Phe163Gly | This paper | Iwasaki lab | |
| Peptide, recombinant protein | H. sapiens eIF4A1 Phe163His | This paper | Iwasaki lab | |
| Peptide, recombinant protein | O. sinensis eIF4A WT | This paper | Iwasaki lab | |
| Peptide, recombinant protein | O. sinensis eIF4A His154Gly | This paper | Iwasaki lab | |
| Peptide, recombinant protein | Ophiocordyceps sp. BRM1 eIF4A iso4 WT | This paper | Iwasaki lab | |
| Peptide, recombinant protein | Ophiocordyceps sp. BRM1 eIF4A iso4 Gly172His | This paper | Iwasaki lab | |
| Peptide, recombinant protein | Ophiocordyceps sp. BRM1 eIF4A iso4 Gly172Phe | This paper | Iwasaki lab | |
| Peptide, recombinant protein | Driselase from Basidiomycetes sp. | Sigma-Aldrich | Cat. #:D9515 | |
| Peptide, recombinant protein | Lysing enzyme from Trichoderma harzianum | Sigma-Aldrich | Cat. #:L1412 | |
| Peptide, recombinant protein | Cas9 nuclease protein NLS | Nippon Gene | Cat. #:316-08651 | |
| Peptide, recombinant protein | Turbo DNase | Thermo Fisher Scientific | Cat. #:AM2238 | |
| Peptide, recombinant protein | RNase I | Lucigen | Cat. #:N6901K | |
| Commercial assay or kit | rRNA depletion by a Ribo-Zero Gold rRNA Removal Kit (Yeast) | Illumina | Cat. #:RZY1324 | |
| Commercial assay or kit | TruSeq Stranded mRNA Kit | Illumina | Cat. #:15027078 | |
| Commercial assay or kit | In-Fusion HD | TaKaRa | Cat. #:639650 | |
| Commercial assay or kit | ProtoScript II Reverse Transcriptase | New England Biolabs | Cat. #:M0368L | |
| Commercial assay or kit | HiFi DNA assembly | New England Biolabs | Cat. #:E2621 | |
| Commercial assay or kit | Ni-NTA agarose | QIAGEN | Cat. #:30230 | |
| Commercial assay or kit | HiTrap Heparin HP column, 1 ml | GE Healthcare | Cat. #:17040601 | |
| Commercial assay or kit | NAP-5 | GE Healthcare | Cat. #:17085302 | |
| Commercial assay or kit | PD-10 | GE Healthcare | Cat. #:17085101 | |
| Commercial assay or kit | Vivaspin 6 (10 kDa MWCO) | Sartorius | Cat. #:VS0601 | |
| Commercial assay or kit | EzStainAQua | ATTO | Cat. #:2332370 | |
| Commercial assay or kit | Black 384-well microplate | Corning | Cat. #:3820 | |
| Commercial assay or kit | T7-Scribe Standard RNA IVT Kit | CELLSCRIPT | Cat. #:C-AS3107 | |
| Commercial assay or kit | ScriptCap m7G Capping System | CELLSCRIPT | Cat. #:C-SCCE0625 | |
| Commercial assay or kit | ScriptCap 2′-O-Methyltransferase Kit | CELLSCRIPT | Cat. #:C-SCMT0625 | |
| Commercial assay or kit | A-Plus Poly(A) Polymerase Tailing Kit | CELLSCRIPT | Cat. #:C-PAP5104H | |
| Commercial assay or kit | Renilla-Glo Luciferase Assay System | Promega | Cat. #:E2720 | |
| Commercial assay or kit | MicroSpin G-25 column | Cytiva | Cat. #:27532501 | |
| Commercial assay or kit | Rabbit Reticulocyte Lysate, Nuclease-Treated | Promega | Cat. #:L4960 | |
| Commercial assay or kit | Potato dextrose agar (PDA) medium | Nissui | Cat. #:05709 | |
| Commercial assay or kit | Potato dextrose broth | BD Biosciences | Cat. #:254920 | |
| Commercial assay or kit | 70 µm cell strainer | Corning | Cat. #:352350 | |
| Commercial assay or kit | Yeast extract | BD Biosciences | Cat. #:212750 | |
| Commercial assay or kit | Filter (0.2 µm pore size) | GE Healthcare | Cat. #:6900-2502 | |
| Commercial assay or kit | 50 ml tube | Falcon, Corning | Cat. #:352070 | |
| Commercial assay or kit | CUGA7 gRNA Synthesis Kit | Nippon Gene | Cat. #:314-08691 | |
| Commercial assay or kit | 50 ml ProteoSave SS tube | Sumitomo Bakelite | Cat. #:631-28111 | |
| Commercial assay or kit | MF membrane (0.45 µm pore size) | Millipore | Cat. #:HAWP04700 | |
| Commercial assay or kit | TRIzol reagent | Thermo Fisher Scientific | Cat. #:15596018 | |
| Commercial assay or kit | Ribo-Minus Eukaryotes Kit for RNA-Seq | Thermo Fisher Scientific | Cat. #:A1083708 | |
| Commercial assay or kit | Direct-zol RNA Microprep Kit | Zymo Research | Cat. #:R2060 | |
| Commercial assay or kit | Illumina Stranded mRNA Prep, Ligation | Illumina | Cat. #:20040532 | |
| Commercial assay or kit | TruSeq Stranded Total RNA Library Prep Gold | Illumina | Cat. #:20020598 | |
| Commercial assay or kit | Vermiculite | VS Kakou | ||
| Commercial assay or kit | Supermix A | Sakata Seed Corp. | Cat. #:72000083 | |
| Commercial assay or kit | Cell strainer (100 µm pore size) | Corning | Cat. #:431752 | |
| Commercial assay or kit | Disposable hemacytometers | Funakoshi | Cat. #:521-10 | |
| Commercial assay or kit | Maxwell RSC Plant RNA Kit | Promega | Cat. #:AS1500 | |
| Commercial assay or kit | ReverTraAce qPCR RT Kit | TOYOBO | Cat. #:FSQ-101 | |
| Commercial assay or kit | THUNDERBIRD Next SYBR qPCR Mix | TOYOBO | Cat. #:QPX-201 | |
| Chemical compound, drug | RocA | Sigma-Aldrich | Cat. #:SML0656 | |
| Chemical compound, drug | Aglafoline | MedChemExpress | Cat. #:HY-19354 | |
| Chemical compound, drug | ADP | Fujifilm Wako Chemicals | Cat. #:019-25091 | |
| Chemical compound, drug | AMP-PNP | Roche | Cat. #:10102547001 | |
| Chemical compound, drug | G418 | Fujifilm Wako Chemicals | Cat. #:078-05961 | |
| Software, algorithm | Trinity | https://github.com/Trinotate/Trinotate/wiki | Grabherr et al., 2011 | |
| Software, algorithm | Trinotate | https://github.com/Trinotate/Trinotate/wiki | Haas et al., 2013 | |
| Software, algorithm | MUSCLE | https://www.ebi.ac.uk/Tools/msa/muscle/ | ||
| Software, algorithm | ESPript 3.0 | http://espript.ibcp.fr/ESPript/ESPript/ | Robert and Gouet, 2014 | |
| Software, algorithm | BLASTp | https://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/ | Camacho et al., 2009 | |
| Software, algorithm | BLASTn | https://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/ | Camacho et al., 2009 | |
| Software, algorithm | MAFFT v7.480 | https://mafft.cbrc.jp/alignment/software/ | Katoh and Standley, 2013 | |
| Software, algorithm | trimAl v1.4.rev15 | https://anaconda.org/bioconda/trimal/files | Capella-Gutiérrez et al., 2009 | |
| Software, algorithm | catfasta2phyml v1.1.0 | https://github.com/nylander/catfasta2phyml | ||
| Software, algorithm | ModelTest-NG v0.1.6 | https://github.com/ddarriba/modeltes | Darriba et al., 2020 | |
| Software, algorithm | RAxML-NG v0.9.0 | https://github.com/amkozlov/raxml-ng | Kozlov et al., 2019 | |
| Software, algorithm | iTOL v6 | https://itol.embl.de/ | Letunic and Bork, 2021 | |
| Software, algorithm | Igor Pro v8.01 | WaveMetrics: https://www.wavemetrics.com/products/igorpro | ||
| Software, algorithm | minimap2 v2.17-r941 | https://anaconda.org/bioconda/minimap2/files | Li, 2018 | |
| Software, algorithm | flye v2.8.1-b1676 | https://anaconda.org/bioconda/flye/files?page=2 | Kolmogorov et al., 2019 | |
| Software, algorithm | nucmer | https://mummer4.github.io/manual/manual.html | Delcher et al., 2003 | |
| Software, algorithm | Fastp v0.21.0 | https://github.com/OpenGene/fastp | Chen et al., 2018 | |
| Software, algorithm | RNAmmer | http://www.cbs.dtu.dk/services/RNAmmer/ | Lagesen et al., 2007 | |
| Software, algorithm | tRNA-scan SE | http://lowelab.ucsc.edu/tRNAscan-SE/ | Chan et al., 2021 | |
| Software, algorithm | STAR v2.7.0a | https://github.com/alexdobin/STAR | Dobin et al., 2013 | |
| Software, algorithm | DESeq2 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html | Love et al., 2014 | |
| Software, algorithm | DAVID | https://david.ncifcrf.gov/home.jsp | Huang et al., 2009a; Huang et al., 2009b | |
| Software, algorithm | StringTie v2.2.1 | https://github.com/gpertea/stringtie | Kovaka et al., 2019 | |
| Other | NGC chromatography system | Bio-Rad | High-performance liquid chromatography | |
| Other | Infinite F-200 PRO | Tecan | Plate reader | |
| Other | EnVision 2104 plate reader | PerkinElmer | Plate reader | |
| Other | GloMax Navigator System | Promega | Cat. #: GM2010 | Microplate luminometer |
| Other | Swinging-bucket rotor | Hitachi | Cat. #:T4SS31 | Centrifuge rotor |
| Other | Centrifuge | Hitachi | Cat. #:CF16RXII | Centrifuge |
| Other | Multi-beads Shocker | YASUI KIKAI | Cat. #:MB2200(S) | Bead mill homogenizer |
| Other | Biotron | NK Systems | Cat. #:LPH-410S and NH350S | Biotron |
| Other | Glass spray | Sansho | Cat. #:81-1192 | Spray |
| Other | Air compressor | NRK Japan | Cat. #:UP-5F | Air compressor |
| Other | 6 mm trepan | Kai Medical | Cat. #:BP-60F | Biopsy Punch |
| Other | 2 ml steel top tube | BMS | Cat. #:MT020-01HS | Sample tube |
| Other | Φ5-mm zirconia beads | Nikkato | Cat. #:5-4060-13 | Zirconia beads |
| Other | Shakemaster NEO | BMS | Cat. #:BMS-mini16 | Bead mill homogenizer |
| Other | Maxwell RSC 48 Instrument | Promega | Cat. #:AS8500 | Automated nucleic acid purification platform |
| Other | MX3000P Real-Time qPCR System | Stratagene | Cat. #:401511 | Real-time qPCR system |
Additional files
-
Supplementary file 1
De novo assembly of the Aglaia-infecting fungus transcriptome.
Summary of de novo-assembled transcripts and genes from Aglaia-infecting fungus RNA-Seq.
- https://cdn.elifesciences.org/articles/81302/elife-81302-supp1-v1.xlsx
-
Supplementary file 2
Top 30 BLASTn hits of the Aglaia-infecting fungus ITS sequence against the NCBI nonredundant nucleotide database.
Nucleotide sequence accessions are listed with subject strain, description, NCBI taxonomy ID, subject accession, and alignment statistics to Aglaia-infecting fungus ITS (percent identity, alignment length, mismatch numbers, gap opens, subject start, subject end, E-value, and bit score).
- https://cdn.elifesciences.org/articles/81302/elife-81302-supp2-v1.xlsx
-
Supplementary file 3
List of fungal species used for the multilocus phylogenetic tree analysis.
Fungal species are listed with host species, strain names, GenBank IDs (ITS, SSU, LSU, TEF1α, and RPB1), and references. The DNA sequences shown in the columns are the best hits from the nucleotide collection searched by BLASTn.
- https://cdn.elifesciences.org/articles/81302/elife-81302-supp3-v1.xlsx
-
Supplementary file 4
List of C. orbiculare strains used in this study.
The C. orbiculare strains used in this study are listed with the strain IDs, genotypes, parental strains, and descriptions.
- https://cdn.elifesciences.org/articles/81302/elife-81302-supp4-v1.xlsx
-
Supplementary file 5
List of oligonucleotides used in this study.
The oligonucleotides used in this study are listed with the sequences, descriptions, and references.
- https://cdn.elifesciences.org/articles/81302/elife-81302-supp5-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/81302/elife-81302-mdarchecklist1-v1.docx