Bacteria: Bringing cells to the edge
Multicellular organisms are, in essence, giant cities of specialized cells. Like the intricate infrastructures that transport people and resources through New York or Amsterdam, vessels and vascular tissues ensure that cells, nutrients and signaling molecules can reach the most remote parts of a plant or an animal. Without these systems, an organism fails to grow and survive.
While bacteria typically live on their own, some can form complex multicellular structures in which cells communicate, coordinate their metabolism and even divide up labor (Tsoi et al., 2018; van Gestel et al., 2015). When cultured on flat surfaces, certain bacterial species create colonies that quickly expand to occupy a large surface area. In fact, these structures become much bigger than what would be possible if newly formed bacteria simply ‘crawled’ from the center of the colony to its outskirts (Warren et al., 2019). For instance, a bacterium within a densely packed group will travel roughly the length of a cell in the time that it takes for the population to double in size (Liu et al., 2021). Many processes have been proposed to aid colony expansion, from mechanical forces to chemically driven bacterial movement (Liu et al., 2021; Sourjik and Wingreen, 2012). Now, in eLife, Yilin Wu, Liang Yang and colleagues – including Ye Li and Shiqi Liu as joint first authors – report a new mechanism that allows cells and molecules to travel long distances at a much faster pace than previously expected (Li et al., 2022).
The team (which is based at Nanyang Technological University, the Chinese University of Hong Kong and the Southern University of Science and Technology) focused on the human pathogen Pseudomonas aeruginosa. These opportunistic bacteria form colonies that exhibit exquisite patterns when cultured on a flat agar surface (Luo et al., 2021). Even with the naked eye, Li et al. could observe centimeter-long ‘valleys’ running from the center to the edge of a colony. Under the microscope, the valleys turned out to be a complex network of canals containing fluid flowing towards the colony’s periphery. Several hundred micron wide and 5–10 micron high, these open channels allowed cells to travel thousand times faster than what they could have done on their own (Meacock et al., 2021). Once at the edge, the cells settle and may help the colony to expand.
Both mobile and non-mobile strains of P. aeruginosa could create the canals, ruling out a role of cell movement in this process. Instead, Li et al. hypothesized that canal formation was driven by a gradient in surface tension, the force that takes place where the atmosphere and the fluids at the surface of the colony meet. This process would be mediated by rhamnolipids, a type of surfactant molecule produced by P. aeruginosa and which decreases surface tension. Indeed, Li et al. found that colonies of rhamnolipid-deficient mutants could not form canals, unless rhamnolipids were injected to artificially establish a surface gradient. Additional experiments showed that rhamnolipids accumulated symmetrically in the center of the colony; it is therefore intriguing why canals only emerge in certain regions.
To explore this question, Li et al. measured flow speeds in colonies before canal formation. This revealed that some regions experienced higher shear rates – meaning that the fluid moved in such a way that it exerted a stronger parallel force onto the cells. The canals developed as fluid flows in the high shear domains carried cells away, a phenomenon known as shear-induced banding (Divoux et al., 2016). A mathematical model accounting for these mechanisms was able to recapture the timing and spatial dynamics of canal formations.
Strikingly, the team found that, in addition to cells, the canals were also transporting outer membrane vesicles (Figure 1). These ‘bag-like’ structures are released by P. aeruginosa and often contain compounds essential for bacterial infection, defense or communication (Schwechheimer and Kuehn, 2015). Being able to quickly transport these vesicles across long distances could therefore help P. aeruginosa to fight off their competitors. Further experiments showed that colonies of the bacteria Staphylococcus aureus quickly died when they encountered P. aeruginosa canals, but survived when they were away from them. These results suggest that canal formation can help P. aeruginosa establish its population by eradicating competing pathogens.
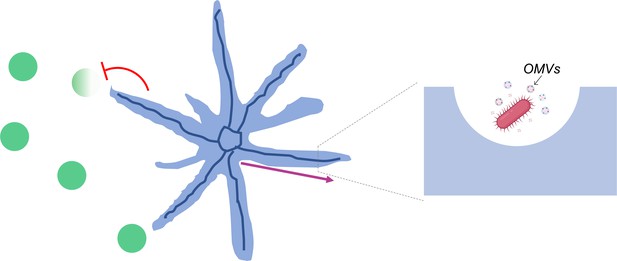
Canals that spontaneously develop in P.aeruginosa colonies enable the long-range directed transport of materials.
Populations of P. aeruginosa bacteria (blue) can adopt a branching pattern under certain conditions. Each branch hosts a single self-organized canal (deep blue) which quickly carries cells (insert; pink) and outer membrane vesicles loaded with molecular cargo (insert; OMVs) from the center to the edge of the colony (pink arrow). These canals help P. aeruginosa eradicate (red flathead arrow) colonies of competing species (green).
It will be intriguing to explore what other physiological functions these structures may have: could they also help bacterial communities to survive antibiotics, lack of resources and other environmental stresses? In particular, fast transportation could enable cells to explore their space more effectively, potentially making a population more competitive by allowing it to expand quickly when nutrients are scarce (Liu et al., 2021).
This study opens the door to many questions that call for future investigation. In the laboratory, canal formation and fluid flow required specific conditions, such as certain levels of humidity. In addition, the canals observed in wildtype colonies were less stable than those in populations of non-mobile mutants (which were used for most of the experiments); in natural settings, the emergence of the channels could easily be disrupted by bacteria being able to move. It therefore remains to be seen how prevalent canals are in the wild, whether they help P. aeruginosa colonize and cause disease in patients, and if they exist in other species.
Even if bacterial canals turn out to be limited in nature, the work of Li et al. opens interesting new avenues for industry. Self-organized bacterial communities can be harnessed to produce valuable chemicals and materials such as biofuels or biodegradable plastics (Kantsler et al., 2020). Being able to modulate how molecules are transported across long distances could allow scientists to better control synthetic communities, for example by loading vesicles with compounds which shape how a colony organizes itself in time and space.
References
-
Shear banding of complex fluidsAnnual Review of Fluid Mechanics 48:81–103.https://doi.org/10.1146/annurev-fluid-122414-034416
-
Pattern engineering of living bacterial colonies using meniscus-driven fluidic channelsACS Synthetic Biology 9:1277–1283.https://doi.org/10.1021/acssynbio.0c00146
-
The spatial organization of microbial communities during range expansionCurrent Opinion in Microbiology 63:109–116.https://doi.org/10.1016/j.mib.2021.07.005
-
Collective colony growth is optimized by branching pattern formation in Pseudomonas aeruginosaMolecular Systems Biology 17:e10089.https://doi.org/10.15252/msb.202010089
-
Bacteria solve the problem of crowding by moving slowlyNature Physics 17:205–210.https://doi.org/10.1038/s41567-020-01070-6
-
Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functionsNature Reviews Microbiology 13:605–619.https://doi.org/10.1038/nrmicro3525
-
Responding to chemical gradients: bacterial chemotaxisCurrent Opinion in Cell Biology 24:262–268.https://doi.org/10.1016/j.ceb.2011.11.008
Article and author information
Author details
Publication history
Copyright
© 2022, Wang and You
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 786
- views
-
- 69
- downloads
-
- 0
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Cell Biology
- Physics of Living Systems
The regulation of cell physiology depends largely upon interactions of functionally distinct proteins and cellular components. These interactions may be transient or long-lived, but often affect protein motion. Measurement of protein dynamics within a cellular environment, particularly while perturbing protein function with small molecules, may enable dissection of key interactions and facilitate drug discovery; however, current approaches are limited by throughput with respect to data acquisition and analysis. As a result, studies using super-resolution imaging are typically drawing conclusions from tens of cells and a few experimental conditions tested. We addressed these limitations by developing a high-throughput single-molecule tracking (htSMT) platform for pharmacologic dissection of protein dynamics in living cells at an unprecedented scale (capable of imaging >106 cells/day and screening >104 compounds). We applied htSMT to measure the cellular dynamics of fluorescently tagged estrogen receptor (ER) and screened a diverse library to identify small molecules that perturbed ER function in real time. With this one experimental modality, we determined the potency, pathway selectivity, target engagement, and mechanism of action for identified hits. Kinetic htSMT experiments were capable of distinguishing between on-target and on-pathway modulators of ER signaling. Integrated pathway analysis recapitulated the network of known ER interaction partners and suggested potentially novel, kinase-mediated regulatory mechanisms. The sensitivity of htSMT revealed a new correlation between ER dynamics and the ability of ER antagonists to suppress cancer cell growth. Therefore, measuring protein motion at scale is a powerful method to investigate dynamic interactions among proteins and may facilitate the identification and characterization of novel therapeutics.
-
- Neuroscience
- Physics of Living Systems
Recent experimental studies showed that electrically coupled neural networks like in mammalian inferior olive nucleus generate synchronized rhythmic activity by the subthreshold sinusoidal-like oscillations of the membrane voltage. Understanding the basic mechanism and its implication of such phenomena in the nervous system bears fundamental importance and requires preemptively the connectome information of a given nervous system. Inspired by these necessities of developing a theoretical and computational model to this end and, however, in the absence of connectome information for the inferior olive nucleus, here we investigated interference phenomena of the subthreshold oscillations in the reference system Caenorhabditis elegans for which the structural anatomical connectome was completely known recently. We evaluated how strongly the sinusoidal wave was transmitted between arbitrary two cells in the model network. The region of cell-pairs that are good at transmitting waves changed according to the wavenumber of the wave, for which we named a wavenumber-dependent transmission map. Also, we unraveled that (1) the transmission of all cell-pairs disappeared beyond a threshold wavenumber, (2) long distance and regular patterned transmission existed in the body-wall muscles part of the model network, and (3) major hub cell-pairs of the transmission were identified for many wavenumber conditions. A theoretical and computational model presented in this study provided fundamental insight for understanding how the multi-path constructive/destructive interference of the subthreshold oscillations propagating on electrically coupled neural networks could generate wavenumber-dependent synchronized rhythmic activity.






