Belly roll, a GPI-anchored Ly6 protein, regulates Drosophila melanogaster escape behaviors by modulating the excitability of nociceptive peptidergic interneurons
Figures
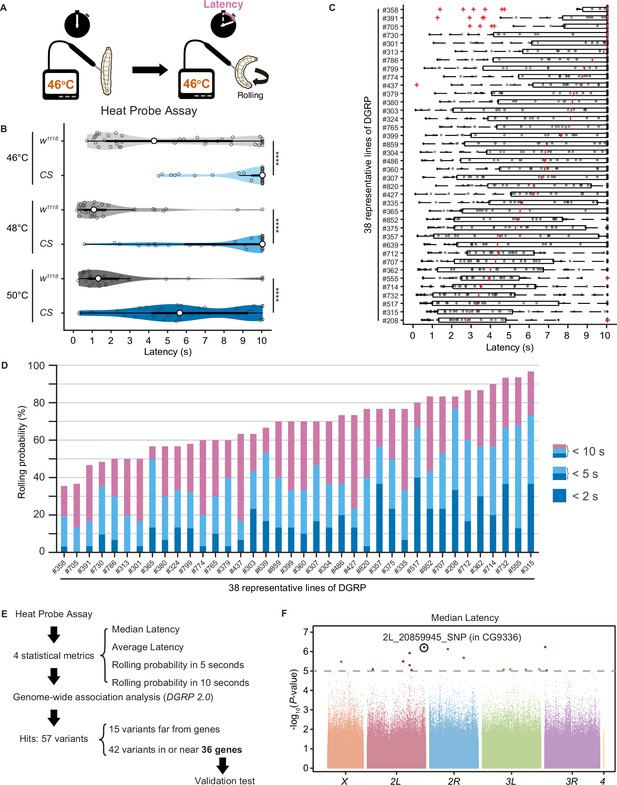
Natural diversity in nociceptive rolling escape behavior in wild-type strains of Drosophila melanogaster.
(A) A schematic representation of the Heat Probe Assay (see ‘Materials and methods’ for details). (B) The latency of rolling escape behavior (rolling latency) is different between two wild-type strains, w1118 and Canton-S (CS) (46°C, [w1118] n = 40, [CS] n = 38; 48°C, [w1118] n = 32, [CS] n = 33; 50°C, [w1118] n = 33, [CS] n = 33; ***p<0.001, Wilcoxon rank-sum test). Violin plots provide a kernel density estimate of the data, where the middle circle shows the median, a boxplot shape indicates 25th and 75th percentiles, and whiskers to the left and right of the box indicate the 90th and 10th percentiles, respectively, in this and the following figures. Each data point represents an individual larva. See source data tables for detailed genotypes in this and the following figures. (C) The rolling latency of 38 representative Drosophila melanogaster Genetic Reference Panel (DGRP) lines in ascending order (n = 30 or 31 larvae/line). Boxplots indicate the median and 25th and 75th percentiles, and whiskers to the left and right of the box indicate the 90th and 10th percentiles, respectively, in this and the following figures. (D) The rolling probability of 38 representative DGRP lines in ascending order (n = 30 or 31 larvae/line). The stacked bar chart indicates the rolling probability within 2, 5, and 10 s, respectively. (E) A diagram showing experimental procedures for the genome-wide association (GWA) analysis. (F) GWA analysis for median rolling latency of the rolling escape behavior. The p-values (−log10 transformed) are shown on the y axis. The gray dotted line marks the nominal p-value threshold (1.0 × 10–5). Each data point corresponds to an individual genetic variant (single-nucleotide polymorphism, deletion, or insertion). Data points are arranged by relative chromosome (genomic) position, the color code indicates the respective chromosome to which they belong. See also Figure 1—source data 1 and Figure 1—figure supplement 1A.
-
Figure 1—source data 1
Summary table of genotypes, statistical testing, and graph data for Figure 1.
- https://cdn.elifesciences.org/articles/83856/elife-83856-fig1-data1-v2.zip
-
Figure 1—source data 2
Genome-wide association results for rolling behavior.
- https://cdn.elifesciences.org/articles/83856/elife-83856-fig1-data2-v2.zip
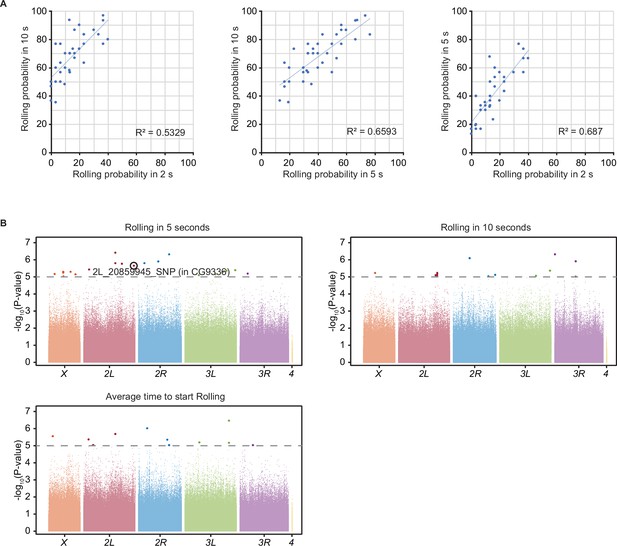
Correlation analysis with rolling probability in three response classes and genome-wide association (GWA) analysis.
(A) A correlation analysis with rolling probability in three response classes (rolling probability in 2, 5, and 10 s) of 38 representative Drosophila melanogaster Genetic Reference Panel (DGRP). Corresponding square of Pearson’s r correlation coefficient (R2) is shown at the bottom right of each graph. (B) GWA analysis for rolling escape behavior, with three distinct statistical metrics (rolling probability in 5 s; rolling probability in 10 s; average rolling latency). The p-values (− log10 transformed) are shown on the y axis. The gray dotted line marks the nominal p-value threshold (1.0 × 10–5). Each data point corresponds to an individual genetic variant (single-nucleotide polymorphism, deletion, or insertion). For each plot, points are arranged by relative chromosome (genomic) position, the color code indicates the respective chromosome to which they belong. See Figure 1—source data 1 for detailed results.
-
Figure 1—figure supplement 1—source data 1
4 Statistical metrics of rolling behavior of Drosophila melanogaster Genetic Reference Panel (DGRP) lines.
- https://cdn.elifesciences.org/articles/83856/elife-83856-fig1-figsupp1-data1-v2.zip
-
Figure 1—figure supplement 1—source data 2
Summary table of genotypes, statistical testing, and graph data for Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/83856/elife-83856-fig1-figsupp1-data2-v2.zip
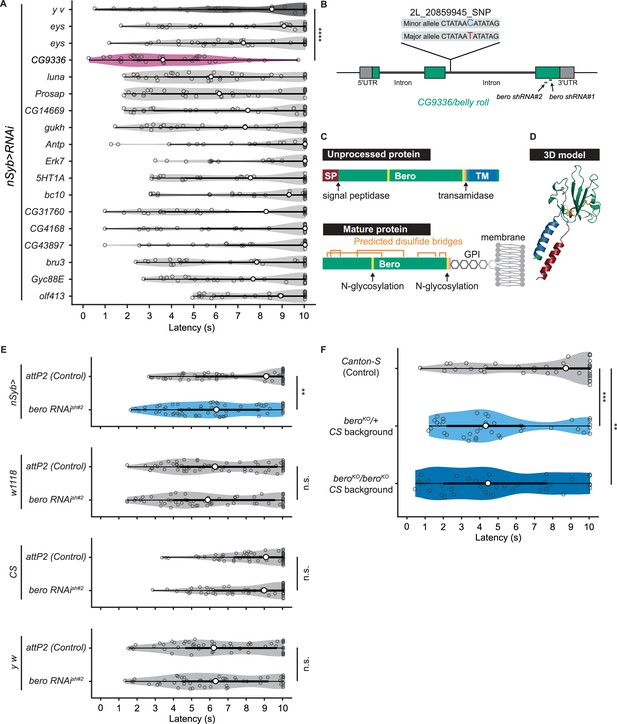
bero negatively regulates nociceptive rolling escape behavior.
(A) A secondary functional screen by pan-neuronal RNA interference. The rolling latency of each UAS-RNAi line was measured. Pan-neuronal knockdown of CG9336/bero reduces rolling latency (Heat Probe Assay, n = 30 or 31 larvae/genotype; ****p<0.0001, Wilcoxon rank-sum test). (B) A schematic representation of bero-associated SNP (2L_20859945_SNP; minor allele, cytosine; major allele, thymine; minor allele frequency = 0.4167). (C) A schematic representation of unprocessed and mature Bero protein. SP, signal peptide; TM, transmembrane region. Orange lines mark the predicted disulfide bonds. Yellow lines mark the predicted N-glycosylation sites. Pink lines mark the predicted GPI-modification site. See also Figure 2—figure supplement 1D. (D) A three-dimensional protein structure prediction of unprocessed Bero protein by AlphaFold2. Red region, signal peptide; blue region, transmembrane region; orange sticks, predicted disulfide bonds; yellow region, predicted N-glycosylation sites; pink region, predicted GPI-modification site. (E) Rolling latency of nSyb>attP2 control (n = 63), pan-neuronal bero knockdown animals (nSyb>bero RNAishRNA#2, n = 65), and the corresponding three sets of effector-controls (w1118 background: attP2 control, n = 64, bero RNAishRNA#2, n = 60; Canton-S background: attP2 control, n = 60, bero RNAishRNA#2, n = 60, y w background: attP2 control, n = 55, bero RNAishRNA#2, n = 55). **p<0.01; n.s., nonsignificant; Wilcoxon rank-sum test. The genetic backgrounds are controlled (see source data tables for detailed genotypes in this and the following figures). (F) Rolling latency of Canton-S control (n = 38), bero heterozygous (beroKO/+, n = 38), and homozygous (beroKO/beroKO, n = 37) mutant animals. **p<0.01, ***p<0.001, Wilcoxon rank-sum test. The bero KO strain had been outcrossed to Canton-S for 11 generations.
-
Figure 2—source data 1
Summary table of genotypes, statistical testing, and graph data for Figure 2.
- https://cdn.elifesciences.org/articles/83856/elife-83856-fig2-data1-v2.zip
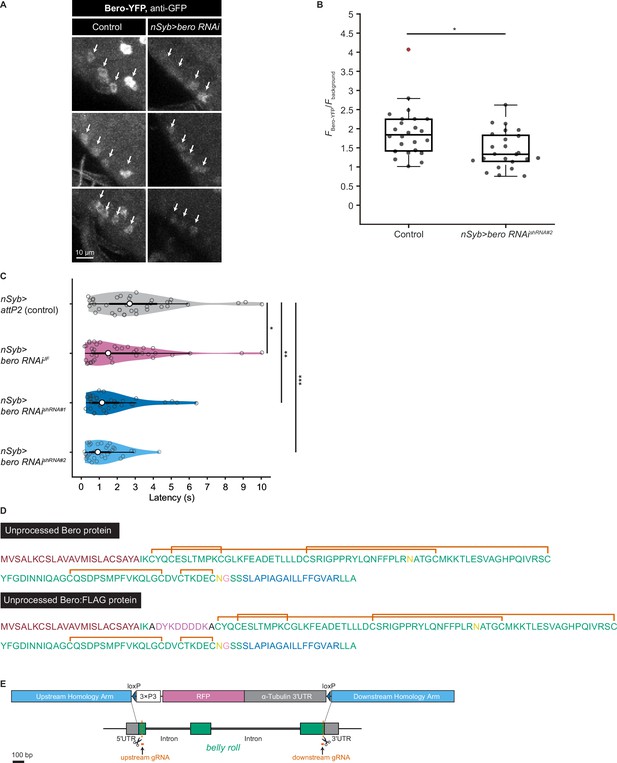
Validation of bero RNAishRNA#2, amino acid sequences of Bero protein, and generation of bero knockout strains.
(A) Representative confocal images showing the expression level of Bero (Bero-YFP, anti-GFP) in ABLK neurons in control and pan-neuronal bero knockdown larvae (nSyb>bero RNAishRNA#2). ABLK neurons are indicated by arrows. Optical setting and analysis process is the same for control and knockdown larvae. (B) A quantitative comparison of normalized fluorescence intensities (FBero-YFP/Fbackground; see ‘Materials and methods’ for details) in ABLK neurons in control (n = 24 neurons from three animals) and pan-neuronal bero knockdown larvae (nSyb>bero RNAishRNA#2, n = 24 neurons from three animals). *p<0.05, unpaired Welch’s t-test. (C) Rolling latency of nSyb>attP2 control (n = 36) and pan-neuronal bero knockdown animals (nSyb>bero RNAiJF, n = 40; nSyb>bero RNAishRNA#1, n = 36; and nSyb>bero RNAishRNA#2, n = 35). *p<0.05, **p<0.01, ***p<0.001, Wilcoxon rank-sum test. (D) Annotated amino acid sequences of unprocessed Bero protein and unprocessed Bero:FLAG protein. The text colors indicate the predicted signal peptide (red), the predicted transmembrane region (blue), the predicted N-glycosylation sites (yellow), the predicted GPI-modification site (pink), and the FLAG-tag sequence (purple). The orange brackets mark the predicted disulfide bonds. (E) A schematic representation of the CRISPR/Cas9-mediated bero knockout by homology-dependent repair (HDR). The procedure generates a 1629 bp deletion in the bero gene, replacing the CDS with a 3xP3-RFP cassette (see ‘Materials and methods’ for details).
-
Figure 2—figure supplement 1—source data 1
Summary table of genotypes, statistical testing, and graph data for Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/83856/elife-83856-fig2-figsupp1-data1-v2.zip

Quantification of bero gene expression.
(A) A scatter plot showing that most of the Drosophila melanogaster Genetic Reference Panel (DGRP) core 38 lines with the bero minor allele (indicated by blue circle) showed higher responsiveness than those with the major allele (indicated by red asterisk) in the Heat Probe Assay. (B) Quantification of bero gene expression in the larval CNS of Canton-S control, bero homozygous (beroKO/beroKO) mutant, two DGRP lines with extremely high rolling latency (DGRP_391, DGRP_705), and two with extremely low rolling latency (DGRP_208, DGRP_315) mRNA levels were normalized to αTub84B expression. The photo of the DNA agarose gel is presented in the top panel. The nucleotide variations of bero-associated SNP in each line are indicated in the bottom panel.
-
Figure 2—figure supplement 2—source data 1
Summary table of genotypes and graph data for Figure 2—figure supplement 2.
- https://cdn.elifesciences.org/articles/83856/elife-83856-fig2-figsupp2-data1-v2.zip
-
Figure 2—figure supplement 2—source data 2
Full raw unedited gel and labeled figure with the uncropped gel for Figure 2—figure supplement 2.
- https://cdn.elifesciences.org/articles/83856/elife-83856-fig2-figsupp2-data2-v2.zip
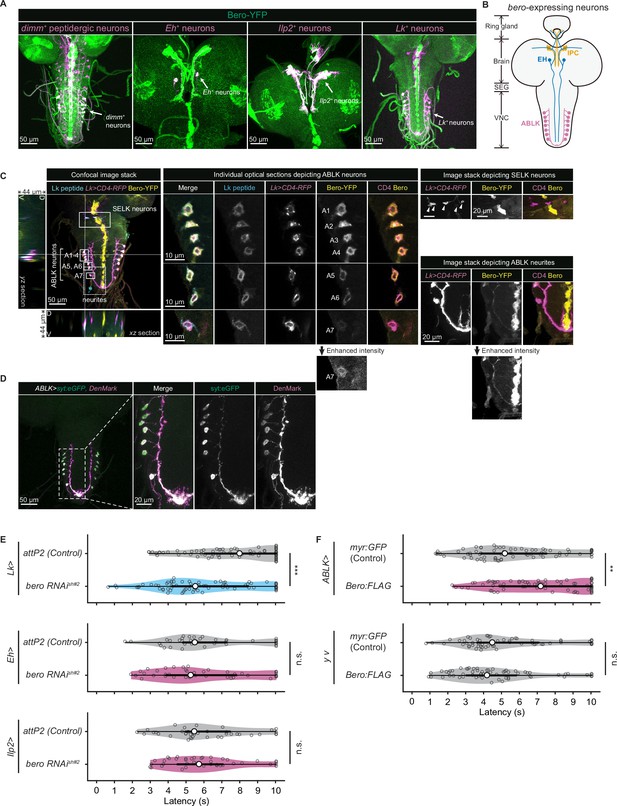
Expression of bero in ABLK neurons plays an essential role in the negative regulation of nociceptive behavior.
(A) Confocal image stacks (maximum projection) showing fluorescence of the endogenous Bero reporter, Bero-YFP (green), and the colocalized peptidergic neurons with GAL4, UAS-CD4-tdTomato (magenta) in third-instar larvae. Scale bars, 50 μm. (B) A schematic representation of bero-expressing neurons. IPC, insulin-producing cells; EH, eclosion hormone-producing neurons; ABLK, abdominal leucokinin-producing neurons; SEG, the subesophageal ganglion; VNC, the ventral nerve cord. (C) Confocal image stack showing endogenous Bero expression (labeled by Bero-YFP, yellow), LK neurons (Lk-GAL4.TH, UAS-CD4-tdTomato, magenta), and leucokinin (labeled by anti-Lk, cyan) in a third-instar larva. Bottom and left panels show an XZ and YZ cross-section of the ABLK somatic region (locations of the cross-sections are indicated by horizontal and vertical gray lines in the primary image), respectively. Middle panels show magnified views of the boxed regions: Individual optical sections depicting ABLK neurons (ABLK neurons are indicated by A1–A7 label, and an image of A7 with enhanced intensity is showed), Right: image stack depicting SELK neurons (indicated by arrowheads), and image stack depicting ABLK neurites (an image with enhanced intensity is showed). Scale bars: 50 μm; 10 μm or 20 μm for the magnified view. (D) Confocal image stack showing the dendrite (labeled by ABLK >DenMark, magenta) and axon terminal markers (labeled by ABLK >syt:eGFP, green) in ABLK neurons in a third-instar larva. Scale bars: 50 μm; 20 μm for the magnified view. (E) Rolling latency of LK-specific bero knockdown larvae (Lk>bero RNAishRNA #2, n = 73) and control (Lk>attP2, n = 75). Rolling latency of Eh-specific bero knockdown larvae (Eh>bero RNAishRNA #2, n = 38) and control (Eh>attP2, n = 41). Rolling latency of Ilp2-specific bero knockdown larvae (Ilp2>bero RNAishRNA #2, n = 40) and control (Ilp2>attP2, n = 39). ***p<0.001; n.s., nonsignificant; Wilcoxon rank-sum test. (F) Rolling latency of ABLK-specific bero overexpression larvae (ABLK>Bero:FLAG, n = 72), control (ABLK>myr:GFP, n = 73), and the corresponding effector-controls (y v background: myr:GFP control, n = 60, Bero:FLAG, n = 61). **p<0.01; n.s., nonsignificant; Wilcoxon rank-sum test.
-
Figure 3—source data 1
Summary table of genotypes, statistical testing, and graph data for Figure 3.
- https://cdn.elifesciences.org/articles/83856/elife-83856-fig3-data1-v2.zip

bero knockdown in nociception-related neurons and specific overexpression of Bero:FLAG in ABLK neurons.
(A) Confocal image stacks (maximum projection) showing simultaneous fluorescence labeling of endogenous Bero reporter, Bero-YFP (green), and nociception-related neurons (Class IV neurons, ppk>CD4-tdTomato; basin neurons, R72F11>CD4-tdTomato; goro neurons, R69F06>CD4-tdTomato; magenta) in the third-instar larvae. Scale bars, 50 μm. (B) Rolling latency of control (ClassIV>attP2, n = 44; Basin>attP2, n = 40; Goro>attP2, n = 39) and neuron-specific bero knockdown animals (ClassIV>bero RNAishRNA#2, n = 43; Basin>bero RNAishRNA#2, n = 39; Goro>bero RNAishRNA#2, n = 40). n.s., nonsignificant; Wilcoxon rank-sum test. (C) Confocal image stacks (maximum projection) showing specific overexpression of Bero:FLAG (anti-FLAG) in ABLK neurons in third-instar larvae. Scale bars, 50 μm.
-
Figure 3—figure supplement 1—source data 1
Summary table of genotypes, statistical testing, and graph data for Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/83856/elife-83856-fig3-figsupp1-data1-v2.zip
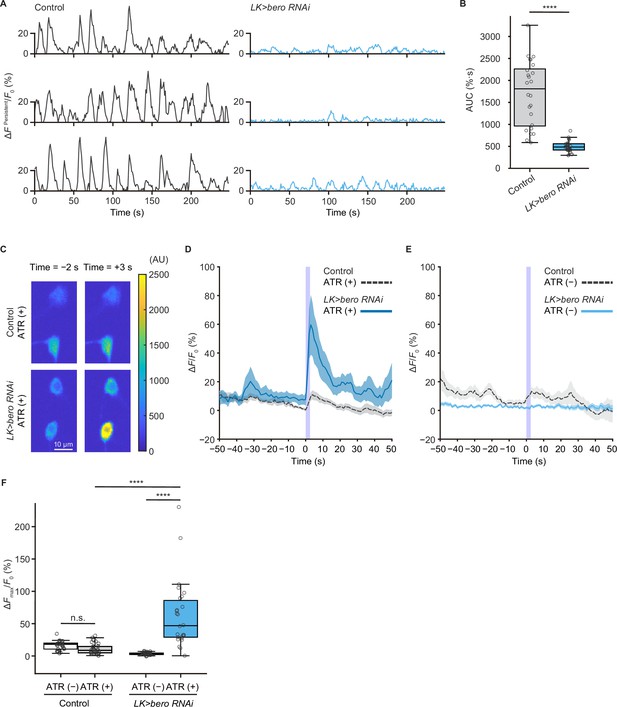
bero is necessary for the maintenance of persistent fluctuating activities and suppression of acute evoked nociceptive activity in ABLK neurons.
(A) Representative results of calcium responses showing the persistent fluctuating calcium signal (∆FPersistent/F0) of ABLK neurons in control (TrpA1>ChR2.T159C, LK>jRCaMP1b) and bero knockdown larvae (TrpA1>ChR2.T159C, LK>jRCaMP1b, bero RNAishRNA #2). (B) A quantitative comparison of area under the curve (AUC) of persistent fluctuating activities of ABLK neurons in control (n = 24 neurons from three animals) and bero knockdown larvae (n = 25 neurons from three animals). ****p<0.0001, Wilcoxon rank-sum test. (C) Representative confocal images of the acute nociceptive responses of ABLK neurons in control (TrpA1>ChR2.T159C, LK>jRCaMP1b) and bero knockdown larvae (TrpA1>ChR2.T159C, LK>jRCaMP1b, bero RNAishRNA #2) after ChR2.T159C-mediated optogenetic activation of Class IV neurons (nociceptors). Scale bars, 10 μm. (D) Time series for calcium responses (∆F/F0; see ‘Materials and methods’ for details) of ABLK neurons in control (TrpA1>ChR2.T159C, LK>jRCaMP1b, ATR (+), n = 29 neurons from three animals) and bero knockdown larvae (TrpA1>ChR2.T159C, LK>jRCaMP1b, bero RNAishRNA #2, ATR (+), n = 27 neurons from three animals) upon optogenetic activation of Class IV neurons (nociceptors). Blue light (470 nm) application is indicated by violet shading beginning at Time 0, and continued for 2.5 s. Light blue and gray shading indicate ± SEM. (E) Time series for calcium responses of ABLK neurons in control (TrpA1>ChR2.T159C, LK>jRCaMP1b, ATR (−), n = 24 neurons from three animals) and bero knockdown larvae (TrpA1>ChR2.T159C, LK>jRCaMP1b, bero RNAishRNA #2, ATR (−), n = 25 neurons from three animals) upon optogenetic activation of Class IV neurons. (F) Quantitative comparison of maximum calcium responses (ΔFmax/F0) of ABLK neurons in control (ATR (−), n = 24 neurons from three animals; ATR (+), n = 29 neurons from three animals) and bero knockdown larvae (ATR (−), n = 25 neurons from three animals; ATR (+), n = 27 neurons from three animals) upon optogenetic activation of Class IV neurons (nociceptors). ****p<0.0001; n.s., nonsignificant; Kruskal−Wallis test followed by Dunn’s test.
-
Figure 4—source data 1
Summary table of genotypes, statistical testing, and graph data for Figure 4.
- https://cdn.elifesciences.org/articles/83856/elife-83856-fig4-data1-v2.zip
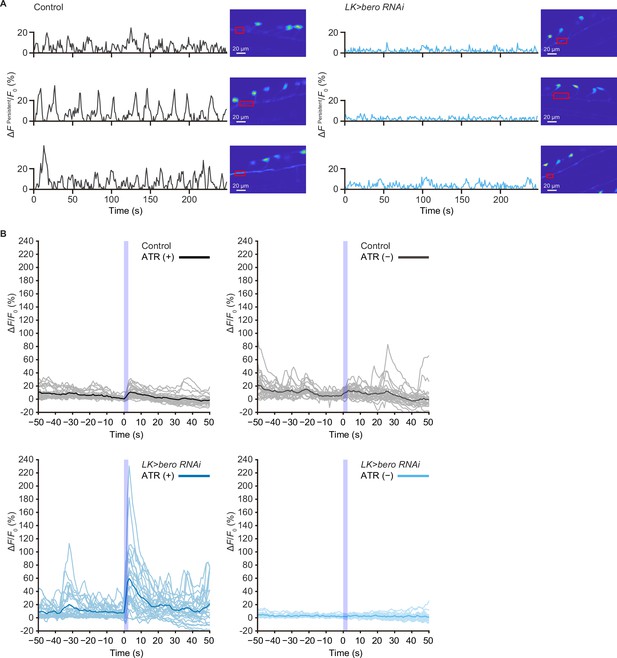
Persistent fluctuating activities at the neurites of ABLK neurons and calcium responses recording traces of control and bero knockdown larvae.
(A) Representative results of calcium responses showing the persistent fluctuating calcium signal (∆FPersistent/F0) at the neurites of ABLK neurons in control (TrpA1>ChR2.T159C, LK>jRCaMP1b) and bero knockdown larvae (TrpA1>ChR2.T159C, LK>jRCaMP1b, bero RNAishRNA #2). Confocal images showing the corresponding region of interest. Scale bars, 20 μm. (B) Time series for calcium responses recording traces (∆F/F0; see ‘Materials and methods’ for details) of ABLK neurons in control (TrpA1>ChR2.T159C, LK>jRCaMP1b, ATR (+), n = 29 neurons from three animals; TrpA1>ChR2.T159C, LK>jRCaMP1b, ATR (−), n = 24 neurons from three animals; gray lines) and bero knockdown larvae (TrpA1>ChR2.T159C, LK>jRCaMP1b, bero RNAishRNA #2, ATR (+), n = 27 neurons from three animals; TrpA1>ChR2.T159C, LK>jRCaMP1b, bero RNAishRNA #2, ATR (−), n = 25 neurons from three animals; light blue lines) upon optogenetic activation of Class IV neurons (nociceptors). Blue light (470 nm) application is indicated by violet shading beginning at Time 0, and continued for 2.5 s. Blue and black lines indicate averaged traces.
-
Figure 4—figure supplement 1—source data 1
Summary table of genotypes for Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/83856/elife-83856-fig4-figsupp1-data1-v2.zip

Knockdown of bero in LK neurons does not affect the free locomotion of larvae.
(A) Temporal color-coded tracking of control (LK>attP2, n = 17) and bero knockdown larvae (LK>bero RNAishRNA #2, n = 22). (B) Average velocity of control (LK>attP2, n = 17) and bero knockdown larvae (LK>bero RNAishRNA #2, n = 22). n.s., nonsignificant; Wilcoxon rank-sum test. (C) Fraction of moving forward frames and head casting frames in control (LK>attP2, n = 17) and bero knockdown larvae (LK>bero RNAishRNA #2, n = 22). n.s., nonsignificant; Wilcoxon rank-sum test.
-
Figure 4—figure supplement 2—source data 1
Summary table of genotypes, statistical testing, and graph data for Figure 4—figure supplement 2.
- https://cdn.elifesciences.org/articles/83856/elife-83856-fig4-figsupp2-data1-v2.zip
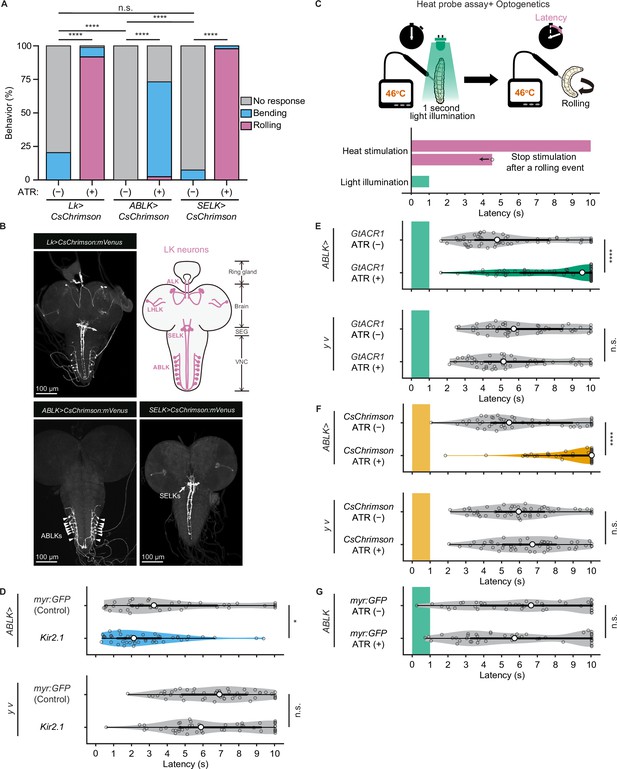
Proper dynamics of acute activity in ABLK neurons is necessary for the control of rolling escape behavior.
(A) Percentage of larvae showing nociceptive escape behavior (rolling and bending) upon optogenetic activation of LK neurons (Lk>CsChrimson, ATR (+), n = 85; ATR (−), n = 49), ABLK neurons (ABLK>CsChrimson, ATR (+), n = 41; ATR (−), n = 22), and SELK neurons (SELK>CsChrimson, ATR (+), n = 46; ATR (−), n = 54). ****p<0.0001; n.s., nonsignificant; Chi-squared test. (B) Confocal image stack showing the labeling of all LK neurons (Lk>CsChrimson.mVenus), ABLK neurons (ABLK>CsChrimson.mVenus), and SELK neurons (SELK>CsChrimson.mVenus) in third-instar larvae. Scale bars: 100 μm. Top-right panel shows a schematic map of LK neuron types. (C) A schematic representation of the Heat Probe Assay combined with optogenetic manipulation (see ‘Materials and methods’ for details). The heat stimulation and light illumination were delivered almost simultaneously: the larva was touched laterally with a heat probe until the initiation of the first 360° rotation of the animal. The response latency was computed as 10 s if it was more than 10 s. A 1-s-long light pulse was delivered for optogenetic manipulation. (D) Rolling latency of ABLK-specific inhibition larvae (ABLK>Kir2.1, n = 45), control larvae (ABLK>myr:GFP, n = 42), and the corresponding effector-controls (y v background: myr:GFP control, n = 50, Kir2.1, n = 50) in the Heat Probe Assay. *p<0.05; n.s., nonsignificant; Wilcoxon rank-sum test. (E) Rolling latency of ABLK-specific optogenetic inhibition larvae (ABLK>GtACR1, ATR (+), n = 68), control larvae (ABLK>GtACR1, ATR (−), n = 54), and the corresponding effector-controls (y v background: GtACR1, ATR (−), n = 50, GtACR1, ATR (+), n = 50) in the Heat Probe Assay. ****p<0.0001; n.s., nonsignificant; Wilcoxon rank-sum test. (F) Rolling latency of ABLK-specific optogenetic activation larvae (ABLK>CsChrimson, ATR (+), n = 53), control larvae (ABLK>CsChrimson, ATR (−), n = 52), and the corresponding effector-controls (y v background: CsChrimson, ATR (−), n = 50, CsChrimson, ATR (+), n = 50) in the Heat Probe Assay. ****p<0.0001; n.s., nonsignificant; Wilcoxon rank-sum test. (G) Rolling latency of the corresponding driver-control larvae (ABLK >myr:GFP, ATR (+), n = 46, ABLK>myr:GFP, ATR (−), n = 45) in the Heat Probe Assay. n.s., nonsignificant; Wilcoxon rank-sum test.
-
Figure 5—source data 1
Summary table of genotypes, statistical testing, and graph data for Figure 5.
- https://cdn.elifesciences.org/articles/83856/elife-83856-fig5-data1-v2.zip
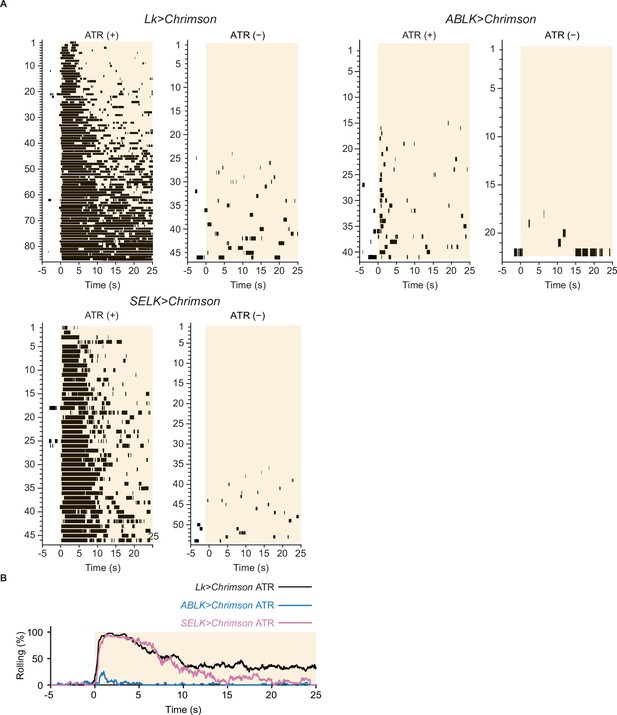
Nociceptive rolling events induced by optogenetic activation.
(A) Raster plots representing nociceptive rolling events induced by LK-specific optogenetic activation (Lk>CsChrimson, ATR (+), n = 85; ATR (−), n = 49), ABLK-specific optogenetic activation (ABLK>CsChrimson, ATR (+), n = 41; ATR (−), n = 22), and SELK-specific optogenetic activation (SELK>CsChrimson, ATR (+), n = 46; ATR (−), n = 54). Orange light (590 nm) application is indicated by orange shading beginning at Time 0 s and continued for 25 s. (B) Time series for the rolling probability of LK-specific (Lk>CsChrimson, ATR (+), n = 85), ABLK-specific (ABLK>CsChrimson, ATR (+), n = 41), and SELK-specific optogenetic activation of larvae (SELK>CsChrimson, ATR (+), n = 46). Orange light (590 nm) application is indicated by the orange shading and continued for 25 s.
-
Figure 5—figure supplement 1—source data 1
Summary table of genotypes and graph data for Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/83856/elife-83856-fig5-figsupp1-data1-v2.zip
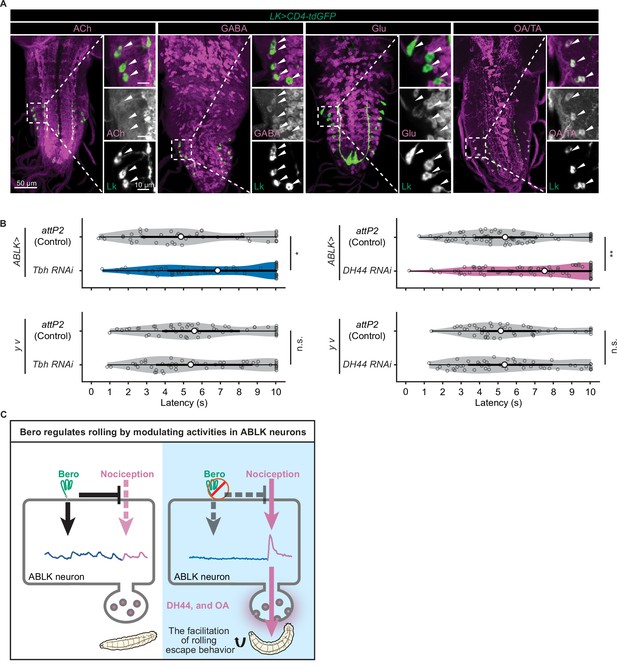
DH44 neuropeptides and octopamine are functional neurotransmitters of ABLK neurons.
(A) Confocal image stacks showing LK neurons (Lk-GAL4.TH, UAS-CD4-tdGFP, green) and distinct types of neurons labeled by the corresponding neurotransmitter markers (cholinergic neurons, anti-ChAT; GABAergic neurons, anti-GABA; glutamatergic neurons, VGlut-T2A-LexA>LexAop-jRCaMP1b; tyraminergic and/or octopaminergic neurons, anti-Tdc2; magenta) in third-instar larvae. Enlarged panels show a magnified view of the boxed region. ABLK neurons are indicated by arrowheads. Scale bars: 50 μm; 10 μm for the magnified view. (B) Rolling latency of ABLK-specific Tbh knockdown larvae (ABLK>Tbh RNAi, n = 59), control larvae (ABLK>attP2, n = 45), and the corresponding effector-controls (y v background: attP2 control, n = 52, Tbh RNAi, n = 51). Rolling latency of ABLK-specific DH44 knockdown larvae (ABLK>DH44 RNAi, n = 83), control larvae (ABLK>attP2, n = 79), and the corresponding effector-controls (y v background: attP2 control, n = 48, DH44 RNAi, n = 50). *p<0.05, **p<0.01; n.s., nonsignificant; Wilcoxon rank-sum test. (C) A model illustrating how Belly roll (Bero) regulates two distinct neuronal activities of a group of bero-expressing neurons, ABLK neurons, which initiate and facilitate the nociceptive escape behavior in Drosophila melanogaster larvae.
-
Figure 6—source data 1
Summary table of genotypes, statistical testing, and graph data for Figure 6.
- https://cdn.elifesciences.org/articles/83856/elife-83856-fig6-data1-v2.zip
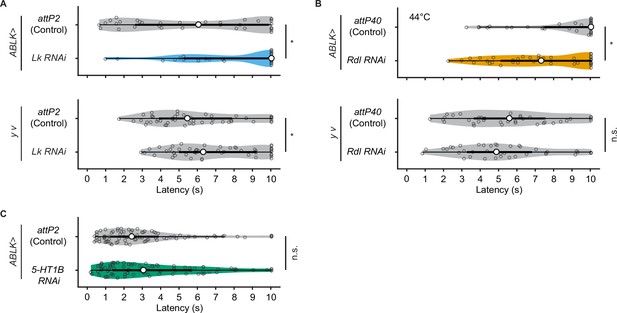
Rolling latency of ABLK-specific Lk, Rdl, or 5-HT1B knockdown larvae.
(A) Rolling latency of ABLK-specific Lk knockdown larvae (ABLK>Lk RNAi, n = 33), control larvae (ABLK>attP2, n = 42), and the corresponding effector-controls (y v background: attP2 control, n = 48, Lk RNAi, n = 50). *p<0.05, Wilcoxon rank-sum test. (B) Rolling latency of ABLK-specific Rdl knockdown larvae (ABLK>Rdl RNAi, n = 38) and control (ABLK>attP40, n = 42) upon lower thermal stimulations (44°C), and the corresponding effector-controls (y v background: attP40 control, n = 38, Rdl RNAi, n = 40). *p<0.05; n.s., nonsignificant; Wilcoxon rank-sum test. (C) Rolling latency of ABLK-specific 5-HT1B knockdown larvae (ABLK>5-HT1B RNAi, n = 83) and control (ABLK>attP2, n = 86). n.s., nonsignificant; Wilcoxon rank-sum test.
-
Figure 6—figure supplement 1—source data 1
Summary table of genotypes, statistical testing, and graph data for Figure 6—figure supplement 1.
- https://cdn.elifesciences.org/articles/83856/elife-83856-fig6-figsupp1-data1-v2.zip

A hypothetical model.
A hypothetical model depicting how Bero may target a receptor that is required for generating the persistent activities, which in turn inhibits the evoked nociceptive responses in ABLK neurons and thereby downregulates the nociceptive rolling behavior. The model also depicts that the persistent activities of the ABLK neurons may reflect certain stresses.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Drosophila melanogaster) | CG9336 | GenBank | FLYB:FBgn0032897 | |
| Strain, strain background (Escherichia coli, DH5α) | E coli | ATCC | ATCC:PTA-1798 | DH5α is an E. coli strain used for general cloning applications |
| Strain, strain background (D. melanogaster) | w[1118] | Bloomington Drosophila Stock Center | BDSC:3605; FLYB:FBst0003605; RRID:BDSC_3605 | |
| Strain, strain background (D. melanogaster) | Canton-S | Drosophila Stocks of Ehime University | DSEU:E-10002 | |
| Strain, strain background (D. melanogaster) | 38 representative DGRP inbred strains | Bloomington Drosophila Stock Center | N/A | See Figure 1—source data 1 in this paper |
| Genetic reagent (D. melanogaster) | nSyb; nSyb-Gal4 | Bloomington Drosophila Stock Center | BDSC:51941; FLYB:FBtp0087725; RRID:BDSC_51941 | FlyBase symbol: P{nSyb-GAL4.P} |
| Genetic reagent (D. melanogaster) | eys; eys RNAi | Bloomington Drosophila Stock Center | BDSC:33764; FLYB:FBti0141009; RRID:BDSC_33764 | FlyBase symbol: P{TRiP.JF02463}attP2 |
| Genetic reagent (D. melanogaster) | eys; eys RNAi | Bloomington Drosophila Stock Center | BDSC:33766; FLYB:FBti0141011; RRID:BDSC_33766 | FlyBase symbol: P{TRiP.JF02708}attP2 |
| Genetic reagent (D. melanogaster) | G9336; bero RNAiJF | Bloomington Drosophila Stock Center | BDSC:31988; FLYB:FBti0130397; RRID:BDSC_31988 | FlyBase symbol: P{TRiP.JF03422}attP2 |
| Genetic reagent (D. melanogaster) | luna; luna RNAi | Bloomington Drosophila Stock Center | BDSC:27084; FLYB:FBti0115451; RRID:BDSC_27084 | FlyBase symbol: P{TRiP.JF02430}attP2 |
| Genetic reagent (D. melanogaster) | Prosap; Prosap RNAi | Bloomington Drosophila Stock Center | BDSC:40929; FLYB:FBti0149838; RRID:BDSC_40929 | FlyBase symbol: P{TRiP.HMS02177}attP2 |
| Genetic reagent (D. melanogaster) | CG14669; CG14669 RNAi | Bloomington Drosophila Stock Center | BDSC:36750; FLYB:FBti0146797; RRID:BDSC_ 36750 | FlyBase symbol: P{TRiP.HMS03010}attP2 |
| Genetic reagent (D. melanogaster) | gukh; gukh RNAi | Bloomington Drosophila Stock Center | BDSC:55858; FLYB:FBti0163218; RRID:BDSC_ 55858 | FlyBase symbol: P{TRiP.HMC03681}attP2 |
| Genetic reagent (D. melanogaster) | Antp; Antp RNAi | Bloomington Drosophila Stock Center | BDSC:64926; FLYB:FBti0184012; RRID:BDSC_ 64926 | FlyBase symbol: P{TRiP.HMC05799}attP2 |
| Genetic reagent (D. melanogaster) | Erk7; Erk7 RNAi | Bloomington Drosophila Stock Center | BDSC:56939; FLYB:FBti0163468; RRID:BDSC_ 56939 | FlyBase symbol: P{TRiP.HMC04378}attP2 |
| Genetic reagent (D. melanogaster) | 5-HT1A; 5-HT1A RNAi | Bloomington Drosophila Stock Center | BDSC:25834; FLYB:FBti0114585; RRID:BDSC_ 25834 | FlyBase symbol: P{TRiP.JF01852}attP2 |
| Genetic reagent (D. melanogaster) | bc10; bc10 RNAi | Bloomington Drosophila Stock Center | BDSC:60486; FLYB:FBti0179272; RRID:BDSC_ 60486 | FlyBase symbol: P{TRiP.HMJ22879}attP40 |
| Genetic reagent (D. melanogaster) | CG31760; CG31760 RNAi | Bloomington Drosophila Stock Center | BDSC:51838; FLYB:FBti0157803; RRID:BDSC_ 51838 | FlyBase symbol: P{TRiP.HMC03410}attP2 |
| Genetic reagent (D. melanogaster) | CG4168; CG4168 RNAi | Bloomington Drosophila Stock Center | BDSC:28736; FLYB:FBti0127300; RRID:BDSC_ 28736 | FlyBase symbol: P{TRiP.JF03163}attP2 |
| Genetic reagent (D. melanogaster) | CG43897; CG43897 RNAi | Bloomington Drosophila Stock Center | BDSC:31560; FLYB:FBti0130595; RRID:BDSC_ 31560 | FlyBase symbol: P{TRiP.JF01132}attP2 |
| Genetic reagent (D. melanogaster) | bru3; bru3 RNAi | Bloomington Drosophila Stock Center | BDSC:43318; FLYB:FBti0151331 RRID:BDSC_ 43318 | FlyBase symbol: P{TRiP.HMS02702}attP40 |
| Genetic reagent (D. melanogaster) | Gyc88E; Gyc88E RNAi | Bloomington Drosophila Stock Center | BDSC:28608; FLYB:FBti0127063; RRID:BDSC_ 28608 | FlyBase symbol: P{TRiP.HM05096}attP2 |
| Genetic reagent (D. melanogaster) | olf413; olf413 RNAi | Bloomington Drosophila Stock Center | BDSC: 29547; FLYB:FBti0128663; RRID:BDSC_ 29547 | FlyBase symbol: P{TRiP.JF02439}attP2 |
| Genetic reagent (D. melanogaster) | bero RNAishRNA#1; | This paper | N/A | See Materials and methods |
| Genetic reagent (D. melanogaster) | bero RNAishRNA#2; | This paper | N/A | See Materials and methods |
| Genetic reagent (D. melanogaster) | nSyb; nSyb-GAL4 | Bloomington Drosophila Stock Center | BDSC:39171; FLYB:FBti0136953; RRID:BDSC_39171 | FlyBase symbol: P{GMR56C10-GAL4}attP2 |
| Genetic reagent (D. melanogaster) | bero-YFP; CG9336(CPTI001654) | Kyoto Drosophila Stock Center | DGRC:115180; FLYB:FBti0143870; RRID:DGGR_115180 | FlyBase symbol: PBac{681.P.FSVS-1}CG9336CPTI001654 |
| Genetic reagent (D. melanogaster) | beroKO; bero knockout | This paper | N/A | See Materials and methods |
| Genetic reagent (D. melanogaster) | UAS-CD4-tdTomato; CD4-tdTomato; CD4-RFP | Bloomington Drosophila Stock Center | BDSC:35837; FLYB:FBti0143424; RRID:BDSC_35837 | FlyBase symbol: PBac{UAS-CD4-tdTom}VK00033 |
| Genetic reagent (D. melanogaster) | R72F11-GAL4; Basin-GAL4; Basin | Bloomington Drosophila Stock Center | BDSC:39786; FLYB:FBti0138028; RRID:BDSC_39786 | FlyBase symbol: P{GMR72F11-GAL4}attP2 |
| Genetic reagent (D. melanogaster) | Ilp2-GAL4.R; Ilp2 | Bloomington Drosophila Stock Center | BDSC:37516; FLYB:FBti0147109 RRID:BDSC_37516 | FlyBase symbol: P{Ilp2-GAL4.R}2 |
| Genetic reagent (D. melanogaster) | Eh.2.4-GAL4; Eh | Bloomington Drosophila Stock Center | BDSC:6301; FLYB:FBti0012534; RRID:BDSC_6301 | FlyBase symbol: P{GAL4-Eh.2.4}C21 |
| Genetic reagent (D. melanogaster) | dimm-GAL4; dimm | Bloomington Drosophila Stock Center | BDSC:25373; FLYB:FBti0004282; RRID:BDSC_ 25373 | FlyBase symbol: P{GawB}dimm929 |
| Genetic reagent (D. melanogaster) | Lk-GAL4; Lk | Bloomington Drosophila Stock Center | BDSC:51993; FLYB:FBti0154847; RRID:BDSC_ 51993 | FlyBase symbol: P{Lk-GAL4.TH}2M |
| Genetic reagent (D. melanogaster) | ppk-GAL4; ppk | Bloomington Drosophila Stock Center | BDSC:32079; FLYB:FBti0131208; RRID:BDSC_ 32079 | FlyBase symbol: P{ppk-GAL4.G}3 |
| Genetic reagent (D. melanogaster) | R69F06-GAL4; Goro-GAL4 | Bloomington Drosophila Stock Center | BDSC:39497; FLYB:FBti0137775; RRID:BDSC_ 39497 | FlyBase symbol: P{GMR69F06-GAL4}attP2 |
| Genetic reagent (D. melanogaster) | tsh-LexA | DOI:10.1080/01677063.2016.1248761 | N/A | J. Simpson, UCSB, Santa Barbara, USA |
| Genetic reagent (D. melanogaster) | Scr-LexA | DOI:10.1080/01677063.2016.1248761 | N/A | J. Simpson, UCSB, Santa Barbara, USA |
| Genetic reagent (D. melanogaster) | tubP-FRT-GAL80-FRT | Bloomington Drosophila Stock Center | BDSC:38881; FLYB:FBti0147582; RRID:BDSC_38881 | FlyBase symbol: P{αTub84B(FRT.GAL80)}3 |
| Genetic reagent (D. melanogaster) | tubP-FRT-GAL80-FRT | Bloomington Drosophila Stock Center | BDSC:38880; FLYB:FBti0147581; RRID:BDSC_38880 | FlyBase symbol: P{αTub84B(FRT.GAL80)}2 |
| Genetic reagent (D. melanogaster) | LexAop-FLP.L | Bloomington Drosophila Stock Center | BDSC:55820; FLYB:FBti0160802; RRID:BDSC_55820 | FlyBase symbol: P{8XLexAop2-FLPL}attP40 |
| Genetic reagent (D. melanogaster) | LexAop-FLP.L | Bloomington Drosophila Stock Center | BDSC:55819; FLYB:FBti0160801; RRID:BDSC_55819 | FlyBase symbol: P{8XLexAop2-FLPL}attP2 |
| Genetic reagent (D. melanogaster) | UAS-Bero:FLAG; Bero:FLAG | This paper | N/A | See Materials and methods |
| Genetic reagent (D. melanogaster) | UAS-myr:GFP; myr:GFP | Bloomington Drosophila Stock Center | BDSC:32197; FLYB:FBti0131941; RRID:BDSC_32197 | FlyBase symbol: P{10XUAS-IVS-myr::GFP}attP2 |
| Genetic reagent (D. melanogaster) | UAS-CsChrimson; CsChrimson; Chrimson | Bloomington Drosophila Stock Center | BDSC:55135; FLYB:FBti0160803; RRID:BDSC_55135 | FlyBase symbol: P{20XUAS-IVS-CsChrimson.mVenus}attP40 |
| Genetic reagent (D. melanogaster) | UAS-CsChrimson; CsChrimson; Chrimson | Bloomington Drosophila Stock Center | BDSC:55136; FLYB:FBti0160804; RRID:BDSC_55136 | FlyBase symbol: P{20XUAS-IVS-CsChrimson.mVenus}attP2 |
| Genetic reagent (D. melanogaster) | UAS-GtACR1-EYFP | DOI: 10.1038/nmeth.4148; Mohammad et al., 2017 | NA | A. Claridge-Chang, Duke-NUS Medical School, Singapore |
| Genetic reagent (D. melanogaster) | UAS-CD4-tdGFP | Bloomington Drosophila Stock Center | BDSC:35839; FLYB:FBti0143426 RRID:BDSC_35839 | FlyBase symbol: P{UAS-CD4-tdGFP}8M2 |
| Genetic reagent (D. melanogaster) | VGlut-LexA | Bloomington Drosophila Stock Center | BDSC:84442; FLYB:FBti0209982; RRID:BDSC_84442 | FlyBase symbol: TI{2A-lexA::GAD}VGlut2A-lexA |
| Genetic reagent (D. melanogaster) | jRCaMP1b; UAS- jRCaMP1b | Bloomington Drosophila Stock Center | BDSC:63793; FLYB:FBti0180189; RRID:BDSC_63793 | FlyBase symbol: PBac{20XUAS-IVS-NES-jRCaMP1b-p10}VK00005 |
| Genetic reagent (D. melanogaster) | jRCaMP1b; UAS- jRCaMP1b | Bloomington Drosophila Stock Center | BDSC:64428; FLYB:FBti0181971; RRID:BDSC_64428 | FlyBase symbol: P{13XLexAop2-IVS-NES-jRCaMP1b-p10}su(Hw)attP5 |
| Genetic reagent (D. melanogaster) | ChR2.T159C | Bloomington Drosophila Stock Center | BDSC:52259; FLYB:FBti0157028; RRID:BDSC_52259 | FlyBase symbol: PBac{10XQUAS-ChR2.T159C-HA}VK00018 |
| Genetic reagent (D. melanogaster) | Lk RNAi; UAS-Lk-RNAi | Bloomington Drosophila Stock Center | BDSC:25798; FLYB:FBti0114549; RRID:BDSC_25798 | FlyBase symbol: P{TRiP.JF01816}attP2 |
| Genetic reagent (D. melanogaster) | DH44 RNAi; UAS-Dh44-RNAi | Bloomington Drosophila Stock Center | BDSC:25804; FLYB:FBti0114555; RRID:BDSC_25804 | FlyBase symbol: P{TRiP.JF01822}attP2 |
| Genetic reagent (D. melanogaster) | Tbh RNAi; UAS-Tbh-RNAi | Bloomington Drosophila Stock Center | BDSC: 27667; FLYB:FBti0128848; RRID:BDSC_27667 | FlyBase symbol: P{TRiP.JF02746}attP2 |
| Genetic reagent (D. melanogaster) | TrpA1-QF | Bloomington Drosophila Stock Center | BDSC:36348; FLYB:FBti0145127; RRID:BDSC_36348 | FlyBase symbol: P{TrpA1-QF.P}attP40 |
| Genetic reagent (D. melanogaster) | attP2 | Bloomington Drosophila Stock Center | BDSC:8622; FLYB:FBti0040535; RRID:BDSC_8622 | FlyBase symbol: P{CaryP}attP2 |
| Genetic reagent (D. melanogaster) | attP40 | Bloomington Drosophila Stock Center | BDSC:36304; FLYB:FBti0114379; RRID:BDSC_36304 | FlyBase symbol: P{CaryP}Msp300attP40 |
| Genetic reagent (D. melanogaster) | UAS-Kir2.1; Kir2.1 | Bloomington Drosophila Stock Center | BDSC:6595; FLYB:FBti0017552; RRID:BDSC_6595 | FlyBase symbol: P{UAS-Hsap\KCNJ2.EGFP}7 |
| Genetic reagent (D. melanogaster) | Rdl RNAi; UAS-Rdl-RNAi | Bloomington Drosophila Stock Center | BDSC:52903; FLYB:FBti0158020; RRID:BDSC_52903 | FlyBase symbol: P{TRiP.HMC03643}attP40 |
| Genetic reagent (D. melanogaster) | 5-HT1B RNAi; UAS-5-HT1B-RNAi | Bloomington Drosophila Stock Center | BDSC:25833; FLYB:FBti0114584; RRID:BDSC_25833 | FlyBase symbol: P{TRiP.JF01851}attP2 |
| Genetic reagent (D. melanogaster) | UAS-DenMark; DenMark | Bloomington Drosophila Stock Center | BDSC:33065; FLYB:FBti0132510; RRID:BDSC_33065 | FlyBase symbol: P{UAS-DenMark}3 |
| Genetic reagent (D. melanogaster) | UAS-syt:eGFP; syt:GFP | Bloomington Drosophila Stock Center | BDSC:33065; FLYB:FBti0026975; RRID:BDSC_33065 | FlyBase symbol: P{UAS-syt.eGFP}3 |
| Antibody | Anti-GFP (chicken polyclonal) | Abcam | Cat#: ab13970; RRID:AB_300798 | IF(1:1000) |
| Antibody | Anti-DN-cadherin (rat monoclonal) | Developmental Studies Hybridoma Bank | Cat#: DN-Ex #8; RRID: AB_528121 | IF(1:100) |
| Antibody | Anti-Lk (rabbit polyclonal) | DOI:10.1016/j.bbrc.2018.03.132; Ohashi and Sakai, 2018 | N/A | IF(1:100) |
| Antibody | Anti-FLAG (mouse monoclonal, M2) | Sigma-Aldrich | Cat#: F3165; RRID:AB_259529 | IF(1:500) |
| Antibody | Anti-ChAT (mouse monoclonal) | Developmental Studies Hybridoma Bank | Cat#: chat4b1; RRID:AB_528122 | IF(1:50) |
| Antibody | Anti-GABA (rabbit polyclonal) | Sigma-Aldrich | Cat#: A2052; RRID: AB_477652 | IF(1:100) |
| Antibody | Anti-Tdc2 (rabbit polyclonal) | Abcam | Cat#: ab128225; RRID:AB_11142389 | IF(1:1000) |
| Antibody | Anti-DYKDDDDK Epitope Tag (rat monoclonal, L5) | Novus Biologicals | Cat#: NBP1-06712; RRID:AB_1625981 | IF(1:500) |
| Antibody | Alexa Fluor 488-conjugated AffiniPure anti-chicken IgY (IgG) (H+L) (donkey polyclonal) | Jackson ImmunoResearch Laboratories Inc | Cat#: 703-545-155; RRID:AB_2340375 | IF(1:500) |
| Antibody | Alexa Fluor 405-conjugated anti-rat IgG (H+L) (goat polyclonal) | Abcam | Cat#: ab175673; RRID:AB_2893021 | IF(1:500) |
| Antibody | Alexa Fluor 405-conjugated anti-rabbit IgG (H+L) (goat polyclonal) | Thermo Fisher Scientific | Cat# :A-31556; RRID:AB_221605 | IF(1:500) |
| Antibody | Alexa Fluor 546-conjugated anti-rat IgG (H+L) (goat polyclonal) | Molecular Probes | Cat#: A-11081; RRID:AB_141738 | IF(1:500) |
| Antibody | Alexa Fluor 546-conjugated anti-mouse IgG (H+L) (goat polyclonal) | Molecular Probes | Cat#: A-11030; RRID:AB_144695 | IF(1:500) |
| Antibody | Alexa Fluor 546-conjugated anti-rabbit IgG (H+L) (goat polyclonal) | Thermo Fisher Scientific | Cat#: A-11035; RRID:AB_2534093 | IF(1:500) |
| Antibody | Alexa Fluor 555-conjugated anti-mouse IgG (H+L) (goat polyclonal) | Thermo Fisher Scientific | Cat#: A-21424; RRID:AB_141780 | IF(1:500) |
| Recombinant DNA reagent | pJFRC7-mCD8::GFP (plasmid) | Addgene | RRID:Addgene_26220 | |
| Recombinant DNA reagent | pJFRC7-FLAG::Bero (plasmid) | This paper | N/A | FLAG::Bero version of pJFRC7-mCD8::GFP; see Materials and methods |
| Recombinant DNA reagent | pVALIUM20 (plasmid) | Drosophila Genomics Resource Center | RRID:DGRC_1467 | |
| Recombinant DNA reagent | pVALIUM20-bero [shRNA#1] (plasmid) | This paper | N/A | See Materials and methods |
| Recombinant DNA reagent | pVALIUM20-bero [shRNA#2] (plasmid) | This paper | N/A | See Materials and methods |
| Recombinant DNA reagent | FI02856 (cDNA) | Drosophila Genomics Resource Center | RRID:DGRC_1621396 | |
| Sequence-based reagent | Primers for bero and αTub84B | This paper | N/A | See Supplementary file 1 |
| Sequence-based reagent | Oligo DNA sequence for bero shRNA#1 and shRNA#2 | This paper | N/A | See Supplementary file 1 |
| Sequence-based reagent | gRNA sequence for bero knockout, see Supplementary file 1 | This paper | N/A | See Supplementary file 1 |
| Sequence-based reagent | Primers for homology arm (bero knockout) | This paper | N/A | See Supplementary file 1 |
| Sequence-based reagent | Primers for validation PCR (bero knockout) | This paper | N/A | See Supplementary file 1 |
| Commercial assay or kit | Sepasol‐RNA I Super G | Nacalai tesque | Cat#: 09379-26 | |
| Commercial assay or kit | RNeasy Mini Kit | QIAGEN | Cat#: 74104 | |
| Commercial assay or kit | ReverTra Ace qPCR RT Master Mix with gDNA Remover | Toyobo | Cat#: FSQ-301 | |
| Commercial assay or kit | KOD-Plus-Neo | Toyobo | Cat#: KOD-401 | |
| Chemical compound, drug | all-trans retinal | Sigma-Aldrich | Cat#: R2500 | |
| Software, algorithm | Arduino IDE 1.8.13 or 2.0.0 | arduino.cc, https://www.arduino.cc/en/software/ReleaseNotes | N/A | |
| Software, algorithm | Fiji | NIH, Bethesda | RRID:SCR_002285 | |
| Software, algorithm | FimTrack | DOI:10.3791/52207; Risse et al., 2017 | N/A | Version 2.1; https://github.com/kostasl/FIMTrack |
| Software, algorithm | MATLAB | The MathWorks, Inc | RRID:SCR_001622 | |
| Software, algorithm | SignalP v. 5.0 | DOI:10.1038/s41587-019-0036-z; Almagro Armenteros et al., 2019 | RRID:SCR_015644 | |
| Software, algorithm | NetNGlyc - 1.0 | DOI:10.1142/9789812799623_0029; Gupta and Brunak, 2001 | RRID:SCR_001570 | |
| Software, algorithm | big-PI Predictor | DOI:10.1006/jmbi.1999.3069; Eisenhaber et al., 1999 | RRID:SCR_001599 | |
| Software, algorithm | TMHMM-2.0 | DOI:10.1006/jmbi.2000.4315; Krogh et al., 2001 | RRID:SCR_014935 | |
| Software, algorithm | AlphaFold2 | DOI:10.1038/s41586-021-03819-2; Jumper et al., 2021 | N/A | Version 2.3.2; https://alphafold.ebi.ac.uk/ |
| Software, algorithm | PyMOL | Schrödinger, Inc | RRID:SCR_000305 |
Additional files
-
Supplementary file 1
Oligonucleotide sequences.
- https://cdn.elifesciences.org/articles/83856/elife-83856-supp1-v2.xlsx
-
Supplementary file 2
Basic sequence analysis of 2L_20859945_SNP neighboring sequence.
- https://cdn.elifesciences.org/articles/83856/elife-83856-supp2-v2.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/83856/elife-83856-mdarchecklist1-v2.pdf





