Functional membrane microdomains and the hydroxamate siderophore transporter ATPase FhuC govern Isd-dependent heme acquisition in Staphylococcus aureus
Figures
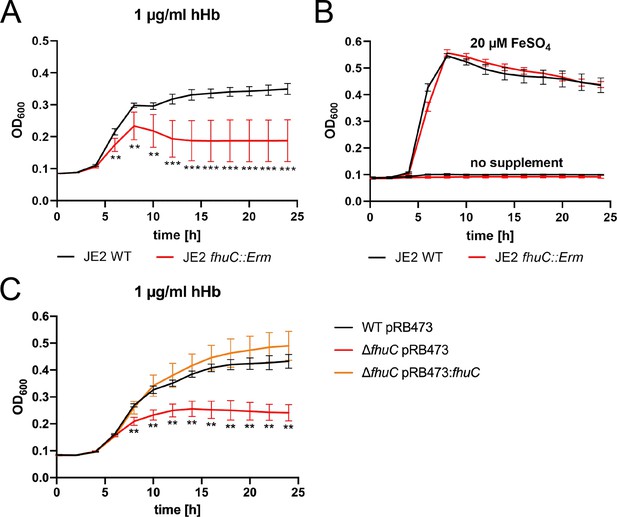
FhuC is needed for hemoglobin (Hb)-dependent proliferation of S. aureus.
One-hundred µl (A, B) or 500 µl (C) of bacterial cultures were grown with human Hb or FeSO4 as the sole source of iron in 96-well (A, B) or 48-well (C) format and OD600 was monitored over time. For reasons of clarity, values taken every 2 hr are displayed. (A, B) S. aureus USA300 JE2 wild type (WT) and USA300 JE2 fhuC::Erm. Means and SD of six experiments are shown. (C) S. aureus Newman ΔfhuC was complemented using a plasmid expressing FhuC from the native promotor (pRB473:fhuC). Newman WT pRB473, ΔfhuC pRB473, and ΔfhuC pRB473:fhuC. Means and SD of three experiments are shown. (A, C) Statistical analysis comparing the WT strains and the fhuC mutants was performed using GraphPad Prism 9 Student’s unpaired t-test. **p<0.01, ***p<0.001.
-
Figure 1—source data 1
FhuC is needed for hemoglobin (Hb)-dependent proliferation of S. aureus.
- https://cdn.elifesciences.org/articles/85304/elife-85304-fig1-data1-v2.zip
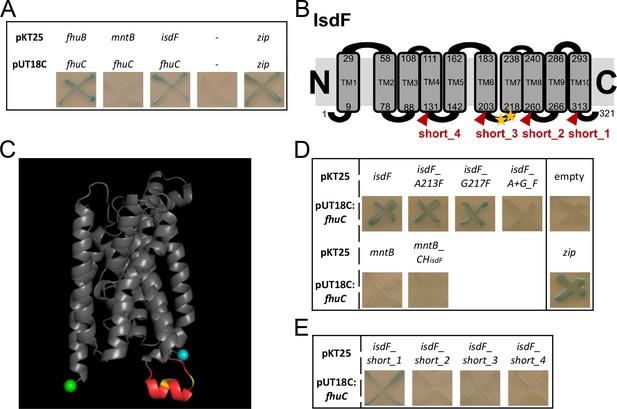
FhuC interacts directly with IsdF.
(A, D, E) Escherichia coli BTH101 was co-transformed with pUT18C:fhuC and pKT25 vectors expressing permeases of interest. Where protein-protein interactions occur, the T25 and T18 catalytic domains of adenylate cyclase dimerize forming an active enzyme which produces cAMP. This activates LacZ expression leading to X-Gal degradation and blue signals on indicator plates. (A) Positive control, leucine zippers (zip). Negative control, empty vectors (pKT25+pUT18C) (-). (B) Schematic representation of the IsdF topology prediction using TOPCONS. The amino acids marking the transmembrane domains (TM) are shown with the conserved A213 and G217 indicated by yellow asterisks. Truncations are indicated in red. (C) IsdF structure prediction using Alphafold with visualization using PyMOL. The coupling helix is shown in red with the conserved A231 and G217 in yellow. The N-terminus is in cyan and the C-terminus in green. (D) Bacterial adenylate cyclase two-hybrid (BACTH) analysis of FhuC and IsdF with single and double amino acid substitutions as well as with the MntB_CHisdF fusion protein (coupling helix IsdF). (E) BACTH analysis of FhuC and truncated IsdF derivatives.
-
Figure 2—source data 1
FhuC interacts directly with IsdF.
- https://cdn.elifesciences.org/articles/85304/elife-85304-fig2-data1-v2.zip
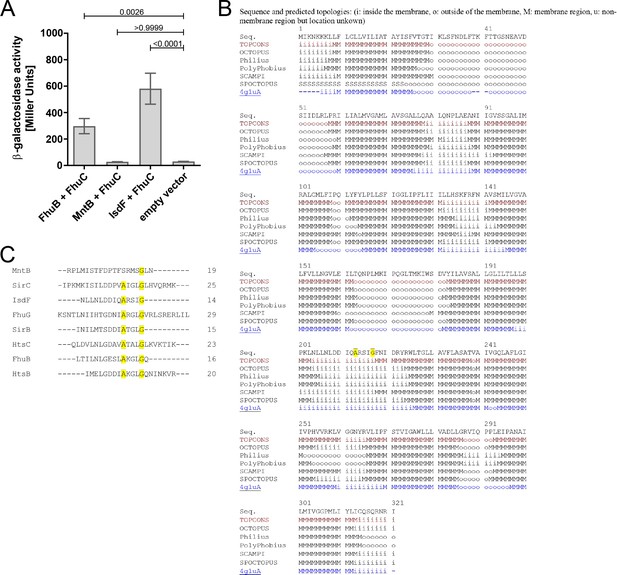
FhuC interaction with iron permeases and topology predictions.
(A) E. coli BTH101 co-transformed with pUT18C:fhuC and pKT25 vectors expressing permeases of interest. Protein-protein interactions leads to β-galactosidase activity. Negative control, empty vectors pKT25+pUT18C. Means and SD of three independent experiments are shown. Statistical analysis, one-way ANOVA followed by Dunett’s test for multiple comparisons was performed using GraphPad Prism 9. (B) TOPCONS modeling of IsdF topology with conserved alanine and glycine residues highlighted in yellow. (B) Clustal Omega sequence alignment of predicted coupling helices of FhuB, FhuG, HtsB, HtsC, SirB, SirC, and IsdF. Conserved alanine and glycine residues are highlighted in yellow.
-
Figure 2—figure supplement 1—source data 1
FhuC interaction with iron permeases and topology predictions.
- https://cdn.elifesciences.org/articles/85304/elife-85304-fig2-figsupp1-data1-v2.zip
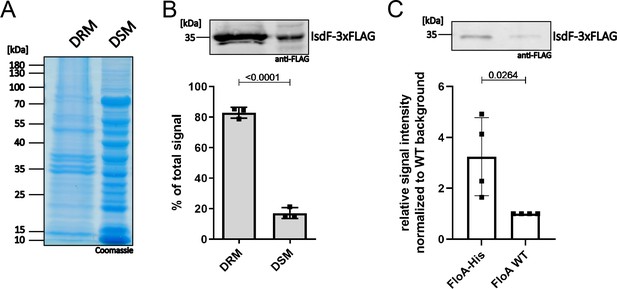
IsdF localizes within functional membrane microdomains (FMMs) and interacts directly with flotillin A (FloA).
S. aureus Newman FloA-His pRB474:isdF-3xFLAG membranes were isolated and (A, B) separated into detergent-resistant membrane (DRM) and detergent-sensitive membrane (DSM) fractions or (C) solubilized with 1% n-dodecyl-β-D-maltopyranosid (DDM) overnight and co-precipitated using Ni-NTA affinity chromatography. (A) Coomassie blue-stained gel of DRM and DSM fractions (equal amounts loaded). (B) Immunoblot analysis and quantification of IsdF in DRM and DSM fractions using anti-FLAG antibody and LI-COR infrared technology. An example of the IsdF-3xFLAG bands in the DRM and DSM fractions is shown. Quantification of signals were calculated as a percentage of the total signal (DRM+DSM). Means and SD of three independent experiments are shown. (C) Co-precipitation analysis using anti-FLAG antibody for detection of IsdF-3xFLAG (pRB474:isdF-3xFLAG) that co-eluted with FloA-His or FloA WT. An example of the IsdF-3xFLAG bands that co-eluted with FloA-His and FloA WT are shown. Quantification of FloA-His IsdF-3xFLAG signals in immunoblots was normalized to FloA WT strain signals (set to 1). Means and SD of four independent experiments are shown. Statistical analysis (Student’s unpaired t-test) was performed using GraphPad Prism 8.
-
Figure 3—source data 1
IsdF localizes within functional membrane microdomains (FMMs) and interacts directly with flotillin A (FloA).
- https://cdn.elifesciences.org/articles/85304/elife-85304-fig3-data1-v2.zip

Input control for co-immunoprecipitation of flotillin A (FloA) and IsdF.
Membranes of S. aureus Newman FloA-His pRB474:isdF-3xFLAG or FloA WT pRB474:isdF-3xFLAG were purified and analyzed prior to Ni-NTA purification. Equal volumes of samples were analyzed by SDS-PAGE and western blotting. Anti-FLAG antibodies were used to detect IsdF-3xFLAG and quantified using LI-COR infrared technology. Signals derived from the FloA WT strain were set to 1 and signals derived from the FloA-His strain are expressed in relation to this value. Means and SD of four independent experiments are shown. Statistical analysis was performed using GraphPad Prism 8 unpaired t-test.
-
Figure 3—figure supplement 1—source data 1
Input control fro co-immunoprecipitation of flotillin A (FloA) and IsdF.
- https://cdn.elifesciences.org/articles/85304/elife-85304-fig3-figsupp1-data1-v2.zip
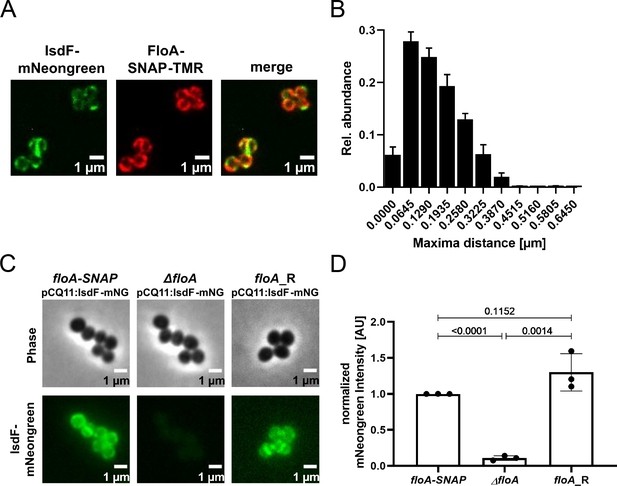
Flotillin A (FloA) is crucial for spatial organization of IsdF in the membrane of S. aureus.
(A) Examples of a fluorescence micrograph of S. aureus Newman floA-SNAP pCQ11:isdF-mNeongreen. Green: IsdF-mNeongreen. Red: FloA-SNAP-TMR. Scale bars, 1 µm. (B) Quantification of the proximity of IsdF-mNeongreen fluorescence maxima and FloA-SNAP-TMR fluorescence maxima. The distance of each FloA-SNAP-TMR maximum to the nearest IsdF-mNeongreen maximum was measured with pixel (px) accuracy (1 px=0.0645 µm). The histogram shows the relative distribution of determined distances. Bars show means and SD of three independent biological replicates. Total number of maxima measured was n≥876 per replicate for each labeled protein. n≥293 cells per replicate. (C) An example of a fluorescence micrograph of S. aureus Newman floA-SNAP pCQ11:isdF-mNeongreen, ΔfloA pCQ11:isdF-mNeongreen, and floA_R pCQ11:isdF-mNeongreen. Scale bars, 1 µm. (D) Quantification of IsdF-Neongreen fluorescence intensity of individual cells. The bar shows the means and SD of three independent biological experiments. n≥241 cells analyzed per strain. Data was normalized to the respective FloA-SNAP replicate mean. Statistical significance was determined using unpaired two-tailed Student‘s t-test with 95% confidence interval.
-
Figure 4—source data 1
Flotillin A (FloA) is crucial for spatial organization of IsdF in the membrane of S. aureus.
- https://cdn.elifesciences.org/articles/85304/elife-85304-fig4-data1-v2.zip
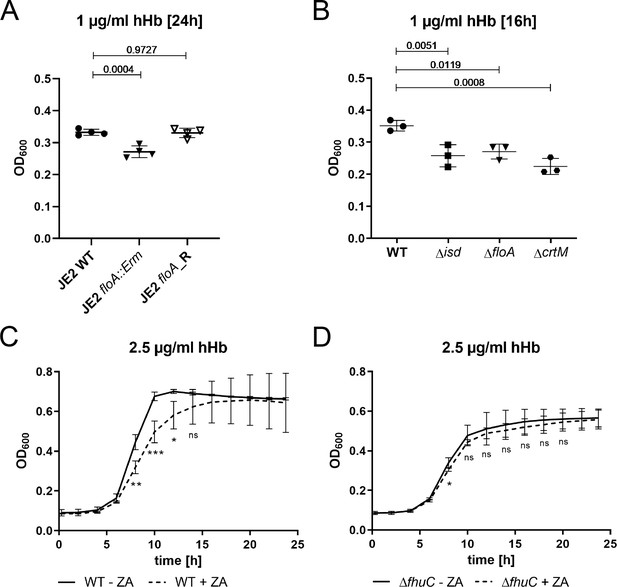
Flotillin A (FloA) and functional membrane microdomains (FMMs) are needed for proliferation with hemoglobin.
(A–D) Strains were grown in iron-limited medium (A: 100 µl in 96-well plates; B–D: 500 µl in 48-well plates). (A) Growth of S. aureus USA300 JE2 WT, ΔfloA::Erm and floA_Revertant (floA_R) in the presence of 1 µg/ml human hemoglobin (hHb). For reasons of clarity, values after 24 hr are displayed. Means and SD of four experiments are shown. (B) Growth of S. aureus Newman WT, Δisd, ΔfloA, and ΔcrtM mutants. Strains were grown in the presence of 1 µg/ml hHb. Values after 16 hr are displayed. Means and SD of three experiments are shown. (C,D) Newman WT (C) and ΔfhuC (D) were grown in the presence of 10 µM zaragozic acid (ZA) and 2.5 µg/ml hHb. Values taken every 2 hr are displayed. Means and SD of four experiments are shown. (A,B) Statistical analysis: Student’s one-way ANOVA followed by Dunett’s test for multiple comparisons was performed using GraphPad Prism 9. (C,D) Statistical analysis: Student’s unpaired t-test was performed using GraphPad Prism 8. *p<0.05, **p<0.01, ***p<0.001.
-
Figure 5—source data 1
Flotillin A (FloA) and functional membrane microdomains (FMMs) are needed for proliferation with hemoglobin.
- https://cdn.elifesciences.org/articles/85304/elife-85304-fig5-data1-v2.zip
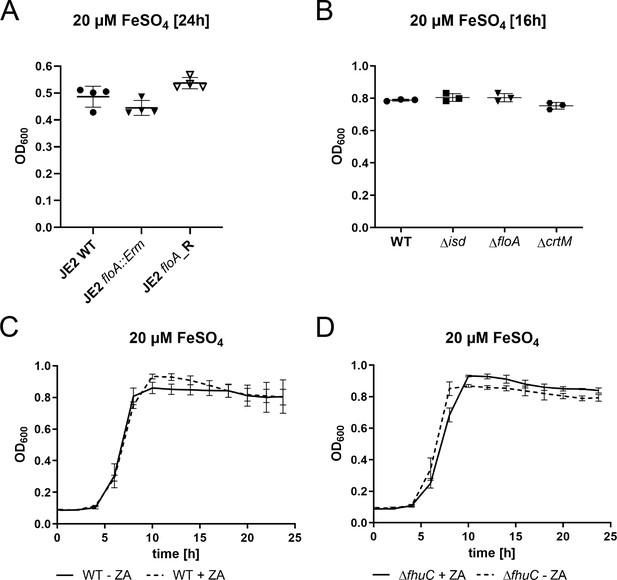
FeSO4 growth controls of floA and functional membrane microdomain (FMM)-deficient mutants.
(A–D) Strains were grown in iron-limited medium (A: 100 µl in 96-well plates; B–D: 500 µl in 48-well plates) in the presence of 20 µM FeSO4. Cultures were inoculated to an OD600=0.005 and OD600 was monitored every 15 min at 37°C orbital shaking using an Epoch2 plate reader. (A) Growth of S. aureus USA300 JE2 WT, ΔfloA::Erm, and floA_Revertant (floA_R). For reasons of clarity, values after 24 hr are displayed. Means and SD of four experiments are shown. (B) Growth of S. aureus Newman WT, Δisd, ΔfloA, and ΔcrtM mutants. Values after 16 hr are displayed. Means and SD of three experiments are shown. (C,D) Newman WT (C) and ΔfhuC (D) were grown in the presence of 10 µM zaragozic acid (ZA) and FeSO4. Values taken every 2 hr are displayed. Means and SD of four experiments are shown.
-
Figure 5—figure supplement 1—source data 1
FeSO4 growth controls of floA and functional membrane microdomain (FMM)-deficient mutants.
- https://cdn.elifesciences.org/articles/85304/elife-85304-fig5-figsupp1-data1-v2.zip
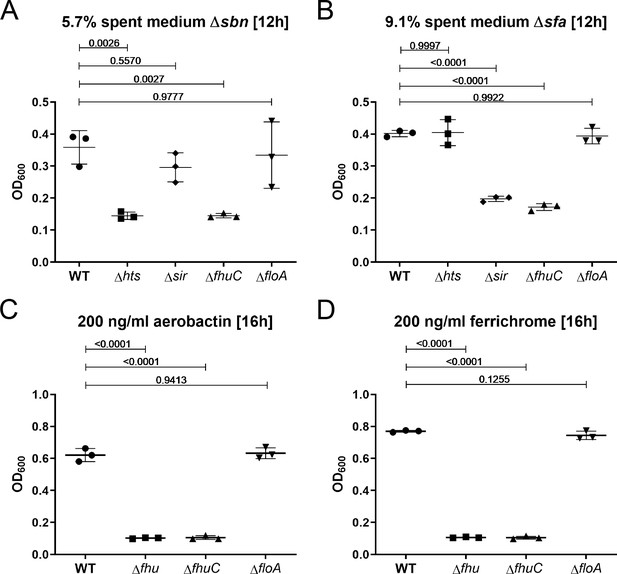
Growth using siderophores is independent of functional membrane microdomains (FMMs).
Newman WT, ΔhtsABC, ΔsirABC, ΔfhuCBG, ΔfhuC, and ΔfloA were grown in the presence of 5.7% spent medium of S. aureus USA300 JE2Δsbn (containing staphyloferrin A) (A), 9.1% spent medium of USA300 JE2 Δsfa (containing staphyloferrin B) (B), 200 ng/ml aerobactin (C) or 200 ng/ml ferrichrome (D). Strains were grown in 500 µl of iron-limited medium in 48-well plates. For reasons of clarity, values after 12 hr (A, B) or 16 hr (C, D) are displayed. Means and SD of three experiments are shown. Statistical analysis: Student’s one-way ANOVA followed by Dunett’s test for multiple comparisons was performed using GraphPad Prism 9.
-
Figure 6—source data 1
Growth using siderophores is independent of functional membrane microdomains (FMMs).
- https://cdn.elifesciences.org/articles/85304/elife-85304-fig6-data1-v2.zip

FeSO4 growth controls of iron transporter and floA mutants.
Newman WT, ΔhtsABC, ΔsirABC, ΔfhuCBG, ΔfhuC, and ΔfloA were grown in the presence 20 µM FeSO4. Strains were inoculated in 500 µl iron-limited medium in 48-well plates. For reasons of clarity, values after 12 hr (A) or 16 hr (B) are displayed. Means and SD of three experiments are shown.
-
Figure 6—figure supplement 1—source data 1
FeSO4 growth controls of iron transporter and floA mutants.
- https://cdn.elifesciences.org/articles/85304/elife-85304-fig6-figsupp1-data1-v2.zip
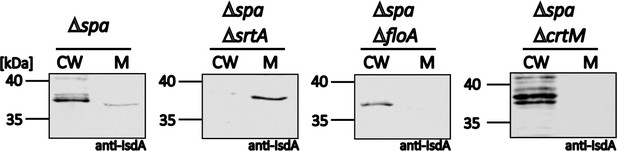
Sortase function does not depend on functional membrane microdomains (FMMs).
S. aureus Newman Δspa, ΔspaΔsrtA::Erm, ΔspaΔfloA::Erm, and ΔspaΔcrtM::Erm cells were grown in iron-limited medium and treated with lysostaphin to gain cell wall (CW) and membrane (M) fractions. Fractions were analyzed by SDS-PAGE and western blotting. IsdA was detected using polyclonal anti-IsdA antibodies.
-
Figure 7—source data 1
Sortase function does not depend on functional membrane microdomains (FMMs).
- https://cdn.elifesciences.org/articles/85304/elife-85304-fig7-data1-v2.zip
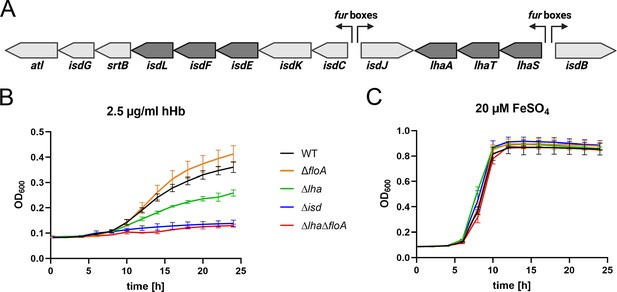
Flotillin A (FloA) is needed for iron-regulated surface determinant (Isd)-dependent proliferation in S. lugdunensis.
(A) Schematic representation of the isd locus in S. lugdunensis N920143. Membrane transporters IsdEFL and LhaSTA in dark gray. Fur boxes are indicated. This figure was created with https://www.biorender.com/. (B, C) S. lugdunensis N920143 WT, ΔfloA, Δlha, Δisd, and ΔlhaΔfloA were grown in 500 µl of iron-limited medium in the presence of 2.5 µg/ml human hemoglobin (hHb) (B) or 20 µM FeSO4 (C) as the sole source of iron in 48-well plates. Values taken every 2 hr are displayed. Means and SD of three experiments are shown.
-
Figure 8—source data 1
Flotillin A (FloA) is needed for iron-regulated surface determinant (Isd)-dependent proliferation in S. lugdunensis.
- https://cdn.elifesciences.org/articles/85304/elife-85304-fig8-data1-v2.zip
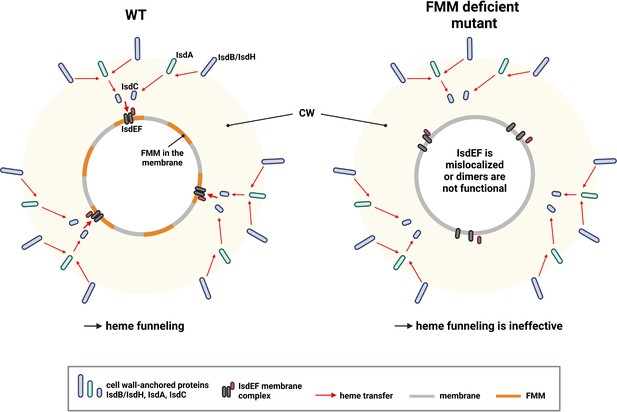
Proposed model of heme funneling over the S. aureus cell envelope.
Cell wall (CW) and membrane of a S. aureus are shown. Heme transfer is indicated by red arrows. The surface-exposed receptors (IsdB and IsdH) extract heme from host hemoproteins and guide it to IsdA and IsdC. We propose that functional membrane microdomains (FMMs) allow structural alignment of IsdC and the membrane receptor IsdEF. Alternatively, the IsdEF complex might be unstable in an FMM or flotillin A (FloA)-deficient strain. This figure was created with https://www.biorender.com/.
-
Figure 9—source data 1
Proposed model of heme funneling over the S. aureus cell envelope.
- https://cdn.elifesciences.org/articles/85304/elife-85304-fig9-data1-v2.zip
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | BTH101 | BACTH System (Euromedex) | Used for BACTH assay; F-, cya-99, araD139, galE15, galK16, rpsL1 (Str r), hsdR2, mcrA1, mcrB1 | |
| Strain, strain background (Escherichia coli) | BTH101 pKT25+pUT18C | This study | BACTH assay | |
| Strain, strain background (Escherichia coli) | BTH101 pKT25:zip+pUT18C:zip | This study | BACTH assay | |
| Strain, strain background (Escherichia coli) | BTH101 pKT25:fhuB+pUT18C:fhuC | This study | BACTH assay | |
| Strain, strain background (Escherichia coli) | BTH101 pKT25:mntB+pUT18C:fhuC | This study | BACTH assay | |
| Strain, strain background (Escherichia coli) | BTH101 pKT25:isdF+pUT18C:fhuC | This study | BACTH assay | |
| Strain, strain background (Escherichia coli) | BTH101 pKT25:isdF_A213F+pUT18C:fhuC | This study | BACTH assay | |
| Strain, strain background (Escherichia coli) | BTH101 pKT25:isdF_G217F+pUT18C:fhuC | This study | BACTH assay | |
| Strain, strain background (Escherichia coli) | BTH101 pKT25:isdF_A+G_F+pUT18C:fhuC | This study | BACTH assay | |
| Strain, strain background (Escherichia coli) | BTH101 pKT25:mntB_CHisdF +pU18C:fhuC | This study | BACTH assay | |
| Strain, strain background (Escherichia coli) | BTH101 pKT25:isdF_short_1+pUT18C:fhuC | This study | BACTH assay | |
| Strain, strain background (Escherichia coli) | BTH101 pKT25:isdF_short_2+pUT18C:fhuC | This study | BACTH assay | |
| Strain, strain background (Escherichia coli) | BTH101 pKT25:isdF_short_3+pUT18C:fhuC | This study | BACTH assay | |
| Strain, strain background (Escherichia coli) | BTH101 pKT25:isdF_short_4+pUT18C:fhuC | This study | BACTH assay | |
| Strain, strain background (Escherichia coli) | SA08B | Monk et al., 2015 | Used for cloning of pIMAY, pRB474 and pRB473 plasmids; DC10BΩPhelp- hsdMS (CC8-2) (SAUSA300_1751) of NRS384 integrated between the atpI and gidB genes | |
| Strain, strain background (Escherichia coli) | SL01B | Heilbronner et al., 2013 | Used for cloning of pIMAY for transformations of S. lugdunensis; DC10B hsdMS+ (S. lugdunensis N920143 (CC1)) | |
| Strain, strain background (Staphylococcus aureus) | USA300 JE2 | Fey et al., 2013 | WT | |
| Strain, strain background (Staphylococcus aureus) | USA300 JE2 Δsbn | This study | Markerless deletion of the entire sbn locus (staphyloferrin B biosynthesis genes; sbnA-I) | |
| Strain, strain background (Staphylococcus aureus) | USA300 JE2 Δsfa | This study | Markerless deletion of the entire sfa locus (staphyloferrin A biosynthesis genes; sfaA-D) | |
| Strain, strain background (Staphylococcus aureus) | USA300 JE2 fhuC::Erm | Fey et al., 2013 | Nebraska transposon library mutant ΔfhuC (SAUSA300_0633) | |
| Strain, strain background (Staphylococcus aureus) | USA300 JE2 floA::Erm | Fey et al., 2013 | Nebraska transposon library mutant ΔfloA (SAUSA300_1533) | |
| Strain, strain background (Staphylococcus aureus) | USA300 JE2 floA_R | This study | Genomic complementation of the Nebraska transposon library mutant floA::Erm with floA | |
| Strain, strain background (Staphylococcus aureus) | COL | Gill et al., 2005 | WT | |
| Strain, strain background (Staphylococcus aureus) | Newman | Lorenz and Duthie, 1952 | WT | |
| Strain, strain background (Staphylococcus aureus) | Newman Δisd | This study | Markerless deletion mutant of the entire isd locus | |
| Strain, strain background (Staphylococcus aureus) | Newman Δfhu | This study | Markerless deletion mutant of the entire fhu (fhuCBG) locus | |
| Strain, strain background (Staphylococcus aureus) | Newman Δhts | This study | Markerless deletion mutant of the entire hts (htsABC) locus | |
| Strain, strain background (Staphylococcus aureus) | Newman Δsir | This study | Markerless deletion mutant of the entire sir (sirABC) locus | |
| Strain, strain background (Staphylococcus aureus) | Newman ΔfhuC | This study | Markerless deletion mutant of fhuC | |
| Strain, strain background (Staphylococcus aureus) | Newman ΔfloA | This study | Markerless deletion mutant of floA | |
| Strain, strain background (Staphylococcus aureus) | Newman ΔcrtM | Wieland et al., 1994 | Deletion mutant of crtM by insertion of Cm resistance gene | |
| Strain, strain background (Staphylococcus aureus) | Newman WT pRB473 | This study | Empty vector control | |
| Strain, strain background (Staphylococcus aureus) | Newman ΔfhuC pRB473 | This study | Empty vector control | |
| Strain, strain background (Staphylococcus aureus) | Newman ΔfhuC pRB473:fhuC | This study | Expression plasmid for fhuC under its native promotor | |
| Strain, strain background (Staphylococcus aureus) | Newman Δspa | This study | Markerless deletion mutant of spa | |
| Strain, strain background (Staphylococcus aureus) | Newman Δspa srtA::Erm | This study | Phage transduction from the Nebraska transposon library mutant srtA::Erm (SAUSA300_2467) into Newman Δspa | |
| Strain, strain background (Staphylococcus aureus) | Newman Δspa floA::Erm | This study | Phage transduction from the Nebraska transposon library mutant floA::Erm (SAUSA300_1533) into Newman Δspa | |
| Strain, strain background (Staphylococcus aureus) | Newman Δspa crtM::Erm | This study | Phage transduction from the Nebraska transposon library mutant crtM::Erm (SAUSA300_2499) into Newman Δspa | |
| Strain, strain background (Staphylococcus aureus) | Newman WT pRB474:isdF-3xFLAG | This study | Expression plasmid for isdF-3xFLAG | |
| Strain, strain background (Staphylococcus aureus) | Newman floA-6xHis pRB474:isdF-3xFLAG | This study | Insertion of linker+6xHis-tag C-terminally of floA; expression plasmid for isdF-3xFLAG | |
| Strain, strain background (Staphylococcus aureus) | Newman floA-SNAP pCQ11:isdF-mNeongreen | This study | Insertion of SNAP tag C-terminally of floA; expression plasmid for isdF-mNeongreen | |
| Strain, strain background (Staphylococcus aureus) | Newman ΔfloA pCQ11:isdF-mNeongreen | This study | Markerless deletion of floA; expression plasmid for isdF-mNeongreen | |
| Strain, strain background (Staphylococcus aureus) | Newman floA_R pCQ11:isdF-mNeongreen | This study | Genomic complementation of the ΔfloA mutant; expression plasmid for isdF-mNeongreen | |
| Strain, strain background (Staphylococcus lugdunensis) | N920143 | Heilbronner et al., 2011 | WT | |
| Strain, strain background (Staphylococcus lugdunensis) | N920143 Δisd | Zapotoczna et al., 2012 | Markerless deletion mutant of the entire isd locus | |
| Strain, strain background (Staphylococcus lugdunensis) | N920143 Δlha | Jochim et al., 2020 | Markerless deletion mutant of lhaSTA | |
| Strain, strain background (Staphylococcus lugdunensis) | N920143 ΔlhaΔfloA | This study | Markerless deletion mutant of lhaSTA and floA | |
| Biological sample (Human) | Human hemoglobin | Own purification (see Materials and methods) | Sex male | |
| Antibody | Mouse monoclonal anti-FLAG M2 | Sigma | F3165 | WB (1:10,000) |
| Antibody | Polyclonal IRDye 800CW goat anti-mouse IgG secondary antibody | LI-COR | 926-32210 | WB (1:10,000) |
| Antibody | Polyclonal rabbit serum anti-IsdA | Prof. J. Geoghegan | WB (1:5000) | |
| Antibody | Polyclonal IRDye 680RD goat anti-rabbit IgG secondary antibody | LI-COR | 926-68071 | WB (1:10,000) |
| Recombinant DNA reagent | pIMAY (plasmid) | Monk et al., 2012 | E. coli/Staphylococcus thermo- sensitive vector for allelic replacement in S. aureus | |
| Recombinant DNA reagent | pIMAY:Δsbn | This study | Plasmid for the deletion of the entire sbn locus | |
| Recombinant DNA reagent | pIMAY:Δsfa | This study | Plasmid for the deletion of the entire sfa locus | |
| Recombinant DNA reagent | pIMAY:ΔfloA complementation | This study | Plasmid for the genomic reversion of in USA300 JE2 floA::Erm and Newman ΔfloA | |
| Recombinant DNA reagent | pIMAY:Δisd | This study | Plasmid for the deletion of the entire isd locus | |
| Recombinant DNA reagent | pIMAY:Δfhu | This study | Plasmid for the deletion of the entire fhu locus | |
| Recombinant DNA reagent | pIMAY:Δhts | This study | Plasmid for the deletion of the entire hts locus | |
| Recombinant DNA reagent | pIMAY:Δsir | This study | Plasmid for the deletion of the entire sir locus | |
| Recombinant DNA reagent | pIMAY:ΔfhuC | This study | Plasmid for the deletion of fhuC | |
| Recombinant DNA reagent | pIMAY:ΔfloA | This study | Plasmid for the deletion of floA | |
| Recombinant DNA reagent | pIMAY:Δspa | This study | Plasmid for the deletion of spa | |
| Recombinant DNA reagent | pIMAY:ΔfloA | This study | Plasmid for the deletion of floA in S. lugdunensis N920143 | |
| Recombinant DNA reagent | pIMAY:floA-6xHis | This study | Plasmid for the addition of 6xHis C-terminally to floA | |
| Recombinant DNA reagent | pIMAY:floA-SNAP | This study | Plasmid for the addition of SNAP C-terminally to floA | |
| Recombinant DNA reagent | pRB473 (plasmid) | Brückner, 1992 | Expression plasmid without a promotor | |
| Recombinant DNA reagent | pRB473:fhuC | This study | fhuC expressing plasmid under its native promotor (fur box) | |
| Recombinant DNA reagent | pRB474 (plasmid) | Brückner, 1992 | Expression plasmid with constitutive promotor | |
| Recombinant DNA reagent | pRB474:isdF-3xFLAG | This study | isdF-3xFLAG expressing plasmid | |
| Recombinant DNA reagent | pCQ11:snap | Lund et al., 2018 | Used as template for SNAP amplification PCR | |
| Recombinant DNA reagent | pCQ11:gfp | C.Quiblier and B. Berger-Bächi | Backbone for pCQ11:mNeongreen | |
| Recombinant DNA reagent | pCQ11:mNeongreen | This study | Backbone for pCQ11 :isdF-mNeongreen | |
| Recombinant DNA reagent | pLOM-S-mNeongreen-EC18153 | Addgene plasmid | # 137075 | mNeongreen template |
| Recombinant DNA reagent | pCQ11:isdF-mNeongreen | This study | isdF with C-terminal mNeongreen fusion; IPTG inducible | |
| Recombinant DNA reagent | pKT25 (plasmid) | BACTH System (Euromedex) | BACTH assay plasmid, N-terminal T25 fragment | |
| Recombinant DNA reagent | pKT25:fhuB | This study | T25 fragment N-terminally of fhuB | |
| Recombinant DNA reagent | pKT25:isdF | This study | T25 fragment N-terminally of isdF | |
| Recombinant DNA reagent | pKT25:mntB | This study | T25 fragment N-terminally of mntB | |
| Recombinant DNA reagent | pKT25:zip | BACTH System (Euromedex) | T25 fragment N-terminally of zip; positive control | |
| Recombinant DNA reagent | pKT25:isdF_short1 | This study | T25 fragment N-terminally of isdF_short1: truncated C-terminus | |
| Recombinant DNA reagent | pKT25:isdF_short2 | This study | T25 fragment N-terminally of isdF_short2: truncated C-terminus+fourth cytosolic loop | |
| Recombinant DNA reagent | pKT25:isdF_short3 | This study | T25 fragment N-terminally of isdF_short3: truncated C-terminus+third cytosolic loop | |
| Recombinant DNA reagent | pKT25:isdF_short4 | This study | T25 fragment N-terminally of isdF_short4: truncated C-terminus+second cytosolic loop | |
| Recombinant DNA reagent | pKT25:isdF_A213F | This study | T25 fragment N-terminally of isdF: alanine position 213 exchanged to phenylalanine | |
| Recombinant DNA reagent | pKT25:isdF_G217F | This study | T25 fragment N-terminally of isdF: glycine position 217 exchanged to phenylalanine | |
| Recombinant DNA reagent | pKT25:isdF_A+G_F | This study | T25 fragment N-terminally of isdF: alanine (213)+glycine (217) exchanged to phenylalanines | |
| Recombinant DNA reagent | pKT25:mntB_CHisdF | This study | T25 fragment N-terminally of mntB; coupling helix of mntB exchanged to the one from isdF | |
| Recombinant DNA reagent | pUT18C | BACTH System (Euromedex) | BACTH assay plasmid, N-terminal T18 fragment | |
| Recombinant DNA reagent | pUT18C:fhuC | This study | T18 fragment N-terminally of fhuC | |
| Recombinant DNA reagent | pUT18C:zip | BACTH System (Euromedex) | T18 fragment N-terminally of zip; positive control | |
| Recombinant DNA reagent | pRB474:mprFdelCysflag | Slavetinsky et al., 2022 | Used as template for 3xFLAG amplification PCR | |
| Sequence-based reagent | Δsbn_PF-A_SmaI | This study | PCR primer | Primer for ΔsbnA-I fragment using pIMAY; CACCTAAAGATCCCGGGACGTCAGTGGC |
| Sequence-based reagent | Δsbn_PR-B | This study | PCR primer | Primer for ΔsbnA-I fragment using pIMAY; CATAGGTGTTTGCCCTACAGAATCTAAC |
| Sequence-based reagent | Δsbn_PF-C | This study | PCR primer | Primer for ΔsbnA-I fragment using pIMAY; CTGTAGGGCAAACACCTATGTAGTTTTACTGTGATGTTGAGGGAAATA |
| Sequence-based reagent | Δsbn_PR-D_KpnI | This study | PCR primer | Primer for ΔsbnA-I fragment using pIMAY; AAATCAGCAAGGTACCACCAATCAGCC |
| Sequence-based reagent | ΔsfaDABC_PF-A_KpnI | This study | PCR primer | Primer for ΔsfaDABC fragment using pIMAY; GATCGGTACCAGTATCTTTAGTTGATGATTCT |
| Sequence-based reagent | ΔsfaDABC_PR-B | This study | PCR primer | Primer for ΔsfaDABC fragment using pIMAY; TAATATATTTATCAATAAGTCTAAGTTGACA |
| Sequence-based reagent | ΔsfaDABC_PF-C | This study | PCR primer | Primer for ΔsfaDABC fragment using pIMAY; ACTTATTGATAAATATATTATAAGGTTATAGAATTTTATTAATCGT |
| Sequence-based reagent | ΔsfaDABC_PR-D | This study | PCR primer | Primer for ΔsfaDABC fragment using pIMAY; CGGAATTCTTCTATTGGTAGTGTAAGTTGGATCA |
| Sequence-based reagent | ΔfloA_PF-A_SacI | This study | PCR primer | Primer for floA::Erm and ΔfloA complementation fragment using pIMAY (floA_R); CCAAGGAGCTCTCAATATGCATTCTATC |
| Sequence-based reagent | ΔfloA comp._PR-B_SmaI | This study | PCR primer | Primer for floA::Erm and ΔfloA complementation fragment using pIMAY (floA_R); CTTCACCAACCCGGGCGATGATTGTTTC |
| Sequence-based reagent | ΔfloA comp._PF-C_SmaI | This study | PCR primer | Primer for floA::Erm and ΔfloA complementation fragment using pIMAY (floA_R); GAAACAATCATCGCCCGGGTTGGTGAAG |
| Sequence-based reagent | ΔfloA_PR-D_KpnI | This study | PCR primer | Primer for floA::Erm and ΔfloA complementation fragment using pIMAY (floA_R); TTTTCGGTACCAATGTCAGTACGAATC |
| Sequence-based reagent | Δisd_PF-A_KpnI | This study | PCR primer | Primer for ΔisdB-G fragment using pIMAY; TAAAGGGAACAAAAGCTGGGTACCAT GCAGAGGACTTACTTGCGTAAAG |
| Sequence-based reagent | Δisd_PR-B | This study | PCR primer | Primer for ΔisdB-G fragment using pIMAY; TAAATTAACAAATTTTAATTGGCGGATG |
| Sequence-based reagent | Δisd_PF-C | This study | PCR primer | Primer for ΔisdB-G fragment using pIMAY; ATTAAAATTTGTTAATTTAAGAATTTAAA GAGGTTGCAGTACTTGTTATG |
| Sequence-based reagent | Δisd_PR-D_SacI | This study | PCR primer | Primer for ΔisdB-G fragment using pIMAY; CGACTCACTATAGGGCGAATTGGAGCTC TCAATTAAATGCACACCTTCAATTAAAGC |
| Sequence-based reagent | Δfhu_PF-A_SacI | This study | PCR primer | Primer for ΔfhuCBG fragment using pIMAY; AATACCTCGAGCTCAGCACGCCATATG CTTTGCTTTTCTTCGAT |
| Sequence-based reagent | Δfhu_PR-B | This study | PCR primer | Primer for ΔfhuCBG fragment using pIMAY; CATAATTTCCCTACTTTCAATAAAATTCTT |
| Sequence-based reagent | Δfhu_PF-C | This study | PCR primer | Primer for ΔfhuCBG fragment using pIMAY; ATTTTATTGAAAGTAGGGAAATTATG TAGTGTCAATGGACACAACTTATTGCTATG |
| Sequence-based reagent | Δfhu_PR-D_KpnI | This study | PCR primer | Primer for ΔfhuCBG fragment using pIMAY; TGCTTTggTAcCTTCTAATATTTTATCAGGTGTAGG |
| Sequence-based reagent | Δhts_PF-A_SacI | This study | PCR primer | Primer for ΔhtsABC fragment using pIMAY; GCACgagCTCATTCGATGTATATGAAAAATTTAC |
| Sequence-based reagent | Δhts_PR-B | This study | PCR primer | Primer for ΔhtsABC fragment using pIMAY; CATCGTTCCACTCCTTAATATGTATAAC |
| Sequence-based reagent | Δhts_PF-C | This study | PCR primer | Primer for ΔhtsABC fragment using pIMAY; TATACATATTAAGGAGTGGAACGATGTA ACTAACATATGATTAGAGTTTAAAAAAG |
| Sequence-based reagent | Δhts_PR-D_KpnI | This study | PCR primer | Primer for ΔhtsABC fragment using pIMAY; GTCAGGTacCAATTTATCTTTTAAAATAG |
| Sequence-based reagent | Δsir_PF-A_SacI | This study | PCR primer | Primer for ΔsirABC fragment using pIMAY; GTTTTgAgCtCTTGATTTTAGCTATCATTG |
| Sequence-based reagent | Δsir_PR-B | This study | PCR primer | Primer for ΔsirABC fragment using pIMAY; CATTGACTAATTAGCCTCCTTCGTG |
| Sequence-based reagent | Δsir_PF-C | This study | PCR primer | Primer for ΔsirABC fragment using pIMAY; AGGAGGCTAATTAGTCAATGTAACG ATATTATTAAAACAAAATG |
| Sequence-based reagent | Δsir_PR-D_KpnI | This study | PCR primer | Primer for ΔsirABC fragment using pIMAY; CTGATGgtAccAATAAGTCAGTAATATAAATTC |
| Sequence-based reagent | PF-A_ΔfhuC_SacI | This study | PCR primer | Primer for ΔfhuC fragment using pIMAY; AATACCTCGAGCTCAGCACGCCATAT GCTTTGCTTTTCTTCGAT |
| Sequence-based reagent | PR-B_ΔfhuC | This study | PCR primer | Primer for ΔfhuC fragment using pIMAY; CATAATTTCCCTACTTTCAATAAAATTCTT |
| Sequence-based reagent | PF-C_ΔfhuC | This study | PCR primer | Primer for ΔfhuC fragment using pIMAY; TTATTGAAAGTAGGGAAATTAT GTAATTAAGTAAGTTAATAT |
| Sequence-based reagent | PR-D_ΔfhuC_KpnI | This study | PCR primer | Primer for ΔfhuC fragment using pIMAY; ATGGTAAGTTGGGTACCCAATTGTTAA TATAATGAATAACGCAATACCA |
| Sequence-based reagent | ΔfloA_PF-A_SacI | This study | PCR primer | Primer for ΔfloA fragment using pIMAY; CCAAGGAGCTCTCAATATGCATTCTATC |
| Sequence-based reagent | ΔfloA_PR-B | This study | PCR primer | Primer for ΔfloA fragment using pIMAY; AAACATGGTATCGCTCCTTTTAATTAATC |
| Sequence-based reagent | ΔfloA_PF-C | This study | PCR primer | Primer for ΔfloA fragment using pIMAY; AAAGGAGCGATACCATGTTTTAAGT CGAGAGGTGATTAAATG |
| Sequence-based reagent | ΔfloA_PR-D_KpnI | This study | PCR primer | Primer for ΔfloA fragment using pIMAY; TTTTCGGTACCAATGTCAGTACGAATC |
| Sequence-based reagent | Δspa_PF-A_SacI | This study | PCR primer | Primer for Δspa fragment using pIMAY; GAAAGAGCTCTTTTAATTCATATGGATGAC |
| Sequence-based reagent | Δspa_PR-B | This study | PCR primer | Primer for Δspa fragment using pIMAY; CATAATATAACGAATTATGTATTGCAATAC |
| Sequence-based reagent | Δspa_PF-C | This study | PCR primer | Primer for Δspa fragment using pIMAY; ACATAATTCGTTATATTATGTAAAAAC AAACAATACACAACGATAG |
| Sequence-based reagent | Δspa_PR-D_KpnI | This study | PCR primer | Primer for Δspa fragment using pIMAY; CAGGTGGGGTACCAGCGAAACTTATTTCAC |
| Sequence-based reagent | N9_PF-A_ΔfloA_SacI | This study | PCR primer | Primer for ΔfloA (for S. lugdunensis N920143) fragment using pIMAY; CTTTATTGGAGCTCCAGTAATAGGCTTTTTTGGCATAG |
| Sequence-based reagent | N9_PR-B_ΔfloA | This study | PCR primer | Primer for ΔfloA (for S. lugdunensis N920143) fragment using pIMAY; CATTAAATCACTCCTATAAATTAATCTATC |
| Sequence-based reagent | N9_PF-C_ΔfloA | This study | PCR primer | Primer for ΔfloA (for S. lugdunensis N920143) fragment using pIMAY; TTTATAGGAGTGATTTAATGTAATTA AAGGGGTGATGTCATGAAC |
| Sequence-based reagent | N9_PR-D_ΔfloA_KpnI | This study | PCR primer | Primer for ΔfloA (for S. lugdunensis N920143) fragment using pIMAY; CGAACAGGTACCAAA TCATCCATAAGTGTATGTTC |
| Sequence-based reagent | PF-A_FloA-6xHis_SacI | This study | PCR primer | Primer for floA-6xHis fragment including linker+6xHis using pIMAY; ATATTGagCtcC TTGTTGGTGGTGCTGGTGAAGAAAC |
| Sequence-based reagent | PR-B_FloA-6xHis | This study | PCR primer | Primer for floA-6xHis fragment including linker+6xHis using pIMAY; GTGATGGTGATGGTGATGCGATCCT CTATGTTCAGGTGACTCATCATCACTTTG |
| Sequence-based reagent | PF-C_FloA-6xHis | This study | PCR primer | Primer for floA-6xHis fragment including linker+6xHis using pIMAY; CATAGAGGATCGCATCACCATCACCATC ACTAAGTCGAGAGGTGATTAAATGAGTG |
| Sequence-based reagent | PR-D_FloA-6xHis_KpnI | This study | PCR primer | Primer for floA-6xHis fragment including linker+6xHis using pIMAY; TTTTCggTAcC AATGTCAGTACGAATCGTTTTAATATC |
| Sequence-based reagent | PF-A_FloA-SNAP_SacI | This study | PCR primer | Primer for floA-SNAP fragment using pIMAY; ATATTGagCtcCTTGTTGGTGGTGCTGGTGAAGAAAC |
| Sequence-based reagent | PR-B_FloA-SNAP | This study | PCR primer | Primer for floA-SNAP fragment using pIMAY; ATTTCGCAATCTTTGTCCATATGTT CAGGTGACTCATCATCACTTTG |
| Sequence-based reagent | PF-SNAP | This study | PCR primer | Primer for floA-SNAP fragment using pIMAY; ATGGACAAAGATTGCGAAATGAAACG |
| Sequence-based reagent | PR-SNAP | This study | PCR primer | Primer for floA-SNAP fragment using pIMAY; TCATCCCAGACCCGGTTTACCCAG |
| Sequence-based reagent | PF-C_FloA-SNAP | This study | PCR primer | Primer for floA-SNAP fragment using pIMAY; GTAAACCGGGTCTGGGATGAGTCGAGAGGTGATTAAATGAGTG |
| Sequence-based reagent | PR-D_FloA-SNAP_KpnI | This study | PCR primer | Primer for floA-SNAP fragment using pIMAY; TTTTCggTAcCAATGTCAGTACGAATCGTTTTAATATC |
| Sequence-based reagent | mNeon-for (NheI, SmaI) | This study | PCR primer | Primer for isdF-mNeongreen construct; TTATGCTAGCTTAACCCGGGATGGCGTCGAAGG |
| Sequence-based reagent | mNeon-rev (AscI) | This study | PCR primer | Primer for isdF-mNeongreen construct; TATAGGCGCGCCTCAACCTCCTTTATAGAG |
| Sequence-based reagent | isdF-for (NheI) | This study | PCR primer | Primer for isdF-mNeongreen construct; GCTCGGCTAGCATGATGATAAAAAATAAAAAG |
| Sequence-based reagent | isdF-rev (SmaI) | This study | PCR primer | Primer for isdF-mNeongreen construct; AATATCCCGGGGATTCGATTTCGTTGAC |
| Sequence-based reagent | pcq11-seq2-for | This study | PCR primer | Primer for isdF-mNeongreen construct; GTTGACTTTATCTACAAGG |
| Sequence-based reagent | pcq11-seq2-rev | This study | PCR primer | Primer for isdF-mNeongreen construct; TCTCGAAAATAATAGAGGG |
| Sequence-based reagent | PF_furbox + fhuC_SacI | This study | PCR primer | Primer for cloning of fur box+fhuC into pRB473; AAAAGAGCTCTTAGTCAATAAGATTG |
| Sequence-based reagent | PR_furbox + fhuC_HindIII | This study | PCR primer | Primer for cloning of fur box+fhuC into pRB473; ATTAACAAGCTTAATTAAGAATAAGCTCTG |
| Sequence-based reagent | PF_474-RBS-IsdF_PstI | This study | PCR primer | Primer for cloning isdF-3xFLAG into pRB474; ATGCCTGCAGaggaggattatgttATGA TGATAAAAAATAAAAAGAAACTAC |
| Sequence-based reagent | PR_474-IsdF | This study | PCR primer | Primer for cloning isdF-3xFLAG into pRB474; CCGTCATGGTCTTTGTAGTCGAT TCGATTTCGTTGACTTTGAC |
| Sequence-based reagent | PF-3xFLAG | This study | PCR primer | Primer for cloning isdF-3xFLAG into pRB474; GACTACAAAGACCATGACGGTGATTAT |
| Sequence-based reagent | PR-474-3xFLAG_SacI | This study | PCR primer | Primer for cloning isdF-3xFLAG into pRB474; TCTATgagctcTCATTTGTCATCGTCATCCTTg |
| Sequence-based reagent | PF_FhuB_pKT25_PstI | This study | PCR primer | Primer for cloning fhuB into pKT25; AAAACTGCAGTTAACATGACAAATA |
| Sequence-based reagent | PR_FhuB_pKT25_EcoRI | This study | PCR primer | Primer for cloning fhuB into pKT25; TGCGTGAATTCTTTGAACTAATCATAT |
| Sequence-based reagent | PF_isdF_pKT25_PstI | This study | PCR primer | Primer for cloning isdF into pKT25; GGATAAAAAATCTGCAGTTGATATGATGATA |
| Sequence-based reagent | PR_isdF-pKT25_EcoRI | This study | PCR primer | Primer for cloning isdF into pKT25; CACTAAACCAGGAATTCTACCGTTTTAGAT |
| Sequence-based reagent | MntB_KT-PF_PstI | This study | PCR primer | Primer for cloning mntB into pKT25; TAGTCAAAGGCTGCAGATAACATGTTAG |
| Sequence-based reagent | MntB_PR_EcoRI | This study | PCR primer | Primer for cloning mntB into pKT25; CTAATAATAAAGGTACTgAaTTcTcCATG |
| Sequence-based reagent | PF_pKT_SmaI | This study | PCR primer | Primer for cloning mntB into pKT25; GAAAACCCGGGCGTTACCCAACTTAATC |
| Sequence-based reagent | PR_pKT_stop_SmaI | This study | PCR primer | Primer for cloning mntB into pKT25; CCAGCCCGGGCGTTGTAAAACTACGG |
| Sequence-based reagent | PF_IsdF_short_after MCS_KpnI | This study | PCR primer | Primer for cloning isdF_short1/2/3/4 into pKT25; GTAGGGTACCGCCGTAGTTTTACAAC |
| Sequence-based reagent | PR_IsdF_short_1_KpnI | This study | PCR primer | Primer for cloning isdF_short1 into pKT25; TTGAggTaccCAAATTAAGTAAATTAG |
| Sequence-based reagent | PR_IsdF_short_2_KpnI | This study | PCR primer | Primer for cloning isdF_short2 into pKT25; GTAAggtaCCCCAACTAGCTTTCTAAC |
| Sequence-based reagent | PR_IsdF_short_3_KpnI | This study | PCR primer | Primer for cloning isdF_short3 into pKT25; TAGTAAAggtAccTTAGGGGACAATAG |
| Sequence-based reagent | PR_IsdF_short_4_KpnI | This study | PCR primer | Primer for cloning isdF_short4 into pKT25; AGAAgGtacCAATATAATTATTAAAAATGG |
| Sequence-based reagent | PF_KT-IsdF_A213F | This study | PCR primer | Primer for cloning isdF_A213F into pKT25; CGACATACAAtttCGAAGTATCGGTTTTAATATTGATCGTTACAGATGG |
| Sequence-based reagent | PR_KT-IsdF_A213F | This study | PCR primer | Primer for cloning isdF_A213F into pKT25; CCATCTGTAACGATCAATATTAAAACCGATACTTCGaaaTTGTATGTCG |
| Sequence-based reagent | PF_KT-IsdF_G217F | This study | PCR primer | Primer for cloning isdF_G217F into pKT25; CGACATACAAGCGCGAAGTATCttTTTTAATATTGATCGTTACAGATGG |
| Sequence-based reagent | PR_KT-IsdF_G217F | This study | PCR primer | Primer for cloning isdF_G217F into pKT25; CCATCTGTAACGATCAATATTAAAAaaGATACTTCGCGCTTGTATGTCG |
| Sequence-based reagent | PF_KT-IsdF_A+G_F | This study | PCR primer | Primer for cloning isdF_A+G_F into pKT25; CGACATACAAtttCGAAGTATCttTTTTAATATTGATCGTTACAGATGG |
| Sequence-based reagent | PR_KT-IsdF_A+G_F | This study | PCR primer | Primer for cloning isdF_A+G_F into pKT25; CCATCTGTAACGATCAATATTAAAAaaGATACTTCGaaaTTGTATGTCG |
| Sequence-based reagent | MntB_KT-P-F (PstI) | This study | PCR primer | Primer for cloning mntB_CHisdF into pKT25; TAGTCAAAGGCTGCAGATAACATGTTAG |
| Sequence-based reagent | PR_MntB-CHisdF | This study | PCR primer | Primer for cloning mntB_CHisdF into pKT25; ACCGATACTTCGCGCTTGTATGTCGTCTAAATTTAGTAAATTATAGAAAATAATGATTAGAATAAGGACGATTGAACCAATCAC |
| Sequence-based reagent | PF_MntB-CHisdF | This study | PCR primer | Primer for cloning mntB_CHisdF into pKT25; AATTTACTAAATTTAGACGACATACAAGCGCGAAGTATCGGTACGACGTTATTACATTACTTTGTGATGTTGTTACTCTCATTAG |
| Sequence-based reagent | PR_pKT_stop_SmaI | This study | PCR primer | Primer for cloning mntB_CHisdF into pKT25; CCAGCCCGGGCGTTGTAAAACTACGG |
| Sequence-based reagent | PF_FhuC-SalI | This study | PCR primer | Primer for cloning fhuC into pUT18C; AGAAAGAAGTCGACTGAAAGTAGGGAAATTATG |
| Sequence-based reagent | PR_FhuC_EcoRI | This study | PCR primer | Primer for cloning fhuC into pUT18C; TATTGAATTCCTTAATTAAGAATAAGCTCT |
| Commercial assay or kit | BACTH System Kit | Euromedex | Cat. No.: EUK001 | |
| Commercial assay or kit | CelLytic MEM protein extraction Kit | Sigma | CE0050 | |
| Chemical compound, drug | RPMI-1640 medium | Sigma | R6504-10L | |
| Chemical compound, drug | Bacto casamino acids | BD Biosciences | 223050 | |
| Chemical compound, drug | EDDHA | LGC Standards | TRC-E335100-10MG | |
| Chemical compound, drug | Zaragozic acid A trisodium salt | Santa Cruz Biotechnology | SC-302001 | |
| Chemical compound, drug | ONPG | Sigma | N1127 | |
| Chemical compound, drug | DDM | Roth | CN26.1 | |
| Chemical compound, drug | Profinity IMAC Resin Ni-charged | Bio-Rad | 1560135 | |
| Other | Ferrichrome | EMC Microcollections | FCH | See Materials and methods: Growth in iron-limited medium |
| Other | Aerobactin | EMC Microcollections | Fe-AERO | See Materials and methods: Growth in iron-limited medium |
| Other | SNAP-Cell TMR-Star | New England Biolabs | S9105S | Materials and methods: Fluorescence microscopy |





