Structural insight into guanylyl cyclase receptor hijacking of the kinase–Hsp90 regulatory mechanism
Figures
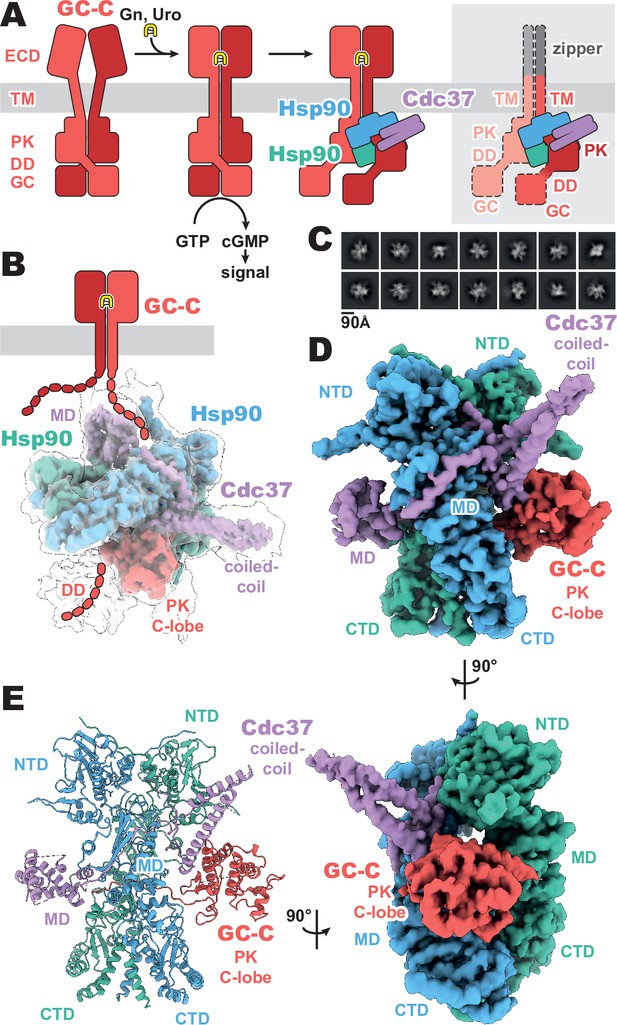
Composition and cryo-EM structure of the GC-C–Hsp90–Cdc37 regulatory complex.
(A) Cartoon representation of the components of guanylyl cyclase C (GC-C) signaling and Hsp90–Cdc37 regulation and the zippered and activated GC-C. GC-C is colored in red, guanylin/uroguanylin (Gn/Uro) in yellow, Hsp90 in blue and teal, and Cdc37 in purple. Extracellular domains (ECD), transmembrane domain (TM), pseudokinase domain (PK), dimerization domain (DD), and guanylyl cyclase domain (GC) are labeled. In the rightmost cartoon, the regions unobserved in the cryo-EM density are in a lighter shade with a dashed outline. (B) The refined and sharpened cryo-EM density map of GC-C–Hsp90–Cdc37, colored as in A, with a transparent overlay of an unsharpened map with additional DD density resolved. Cdc37 coil-coiled and middle domain (MD) are labeled. (C) Reference-free 2D averages for the GC-C–Hsp90–Cdc37 complex. (D) The refined and sharpened cryo-EM density map of GC-C–Hsp90–Cdc37, colored as in A and B, labeled with all domains as in A and B, with the addition of Hsp90 N-terminal domain (NTD), middle domain (MD), and C-terminal domain (CTD). (E) Ribbon representation of a model of GC-C–Hsp90–Cdc37 complex, colored and labeled as in A, B, and C.
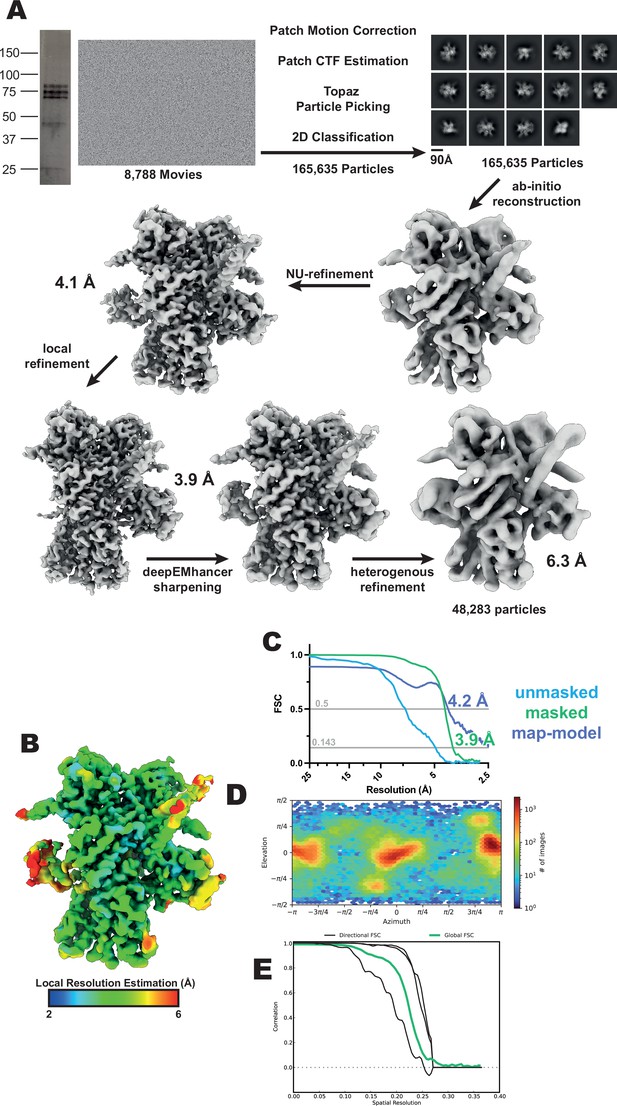
GC-C–Hsp90–Cdc37 complex cryo-EM data processing.
(A) Workflow for cryo-EM data processing. SDS-PAGE gel, representative micrograph, reference-free 2D averages, and cryo-EM maps at the various stages of processing. (B) Local resolution estimation of the finalized cryo-EM map. (C) FSC curve of the reconstruction using gold-standard refinement calculated from unmasked and masked half maps. Map-model FSC curve. (D) Orientational distribution of the reconstruction. (E) Directional FSC curves from 3DFSC (Aiyer et al., 2021).
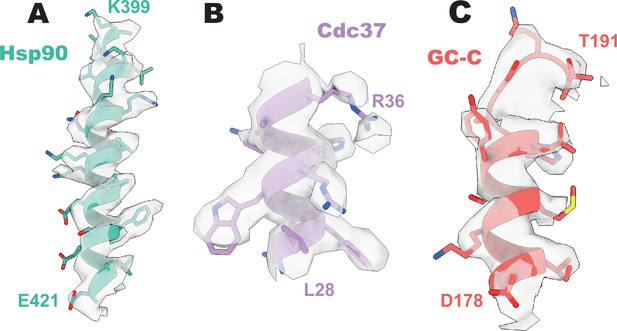
Representative density of GC-C–Hsp90–Cdc37.
(A) Representative density of heat shock protein 90 (Hsp90). (B) Representative density of Cdc37. (C) Representative density of guanylyl cyclase C (GC-C).
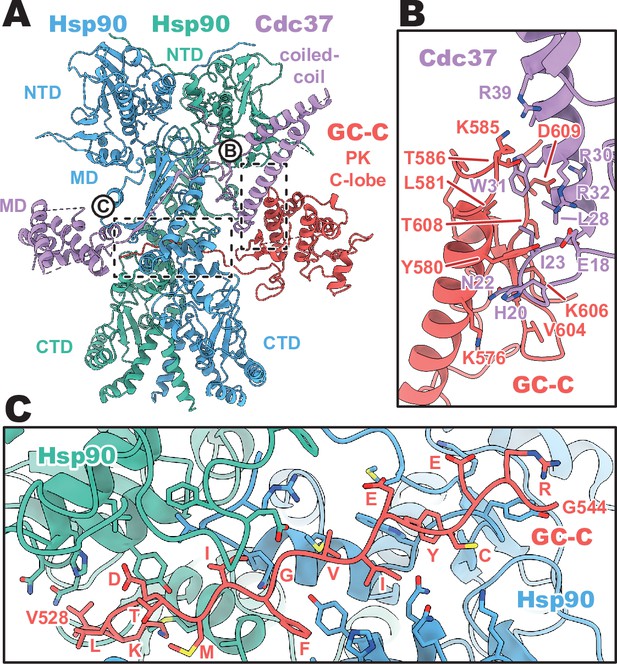
Cdc37 mediated guanylyl cyclase C (GC-C) recruitment and heat shock protein 90 (Hsp90) loading interfaces.
(A) Ribbon representation of a model of GC-C–Hsp90–Cdc37 complex. GC-C is colored in red, Hsp90 in blue and teal, and Cdc37 in purple. Pseudokinase (PK), coil-coiled, middle (MD), C-terminal (CTD), and N-terminal (NTD) domains are labeled. (B) The Cdc37–GC-C interface in ribbon representation, with interacting residues drawn in sticks, colored as in A. (C) The unfolded N-lobe of GC-C PK domain as it passes between the Hsp90 dimer, in ribbon representation, with interacting residues drawn in sticks, colored as in A and B. This region’s sequence is: VKLDTMIFGVIEYCERG.
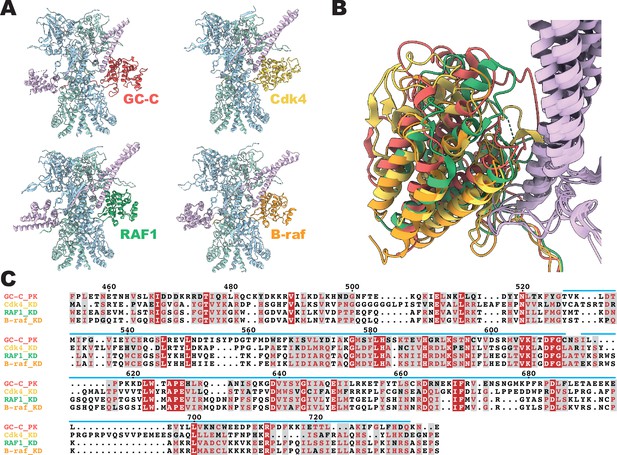
Conservation of Cdc37 mediated heat shock protein 90 (Hsp90) regulation.
(A) Ribbon representation of a model of client–Hsp90β–Cdc37 complexes. Guanylyl cyclase C (GC-C) is colored in red, Cdk4 in yellow (5FWK), RAF1 in green (7Z37), B-raf in orange (7ZR0), Hsp90β in light blue and teal, and Cdc37 in light purple. (B) A structural overlay of the structures in A. (C) A sequence alignment of the pseudokinase domain of GC-C and the kinase domains of Cdk4, RAF1, and B-raf. Sequence numbering per GC-C, with a blue line depicting regions resolved in the cryo-EM density.
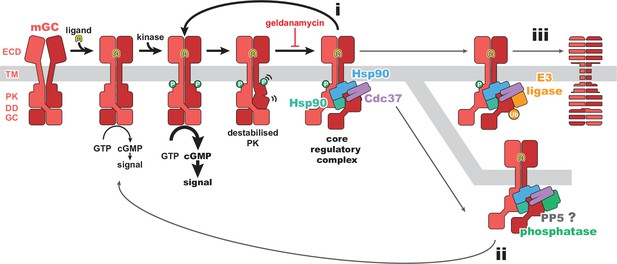
Regulatory mechanisms for membrane receptor guanylyl cyclase (mGC) activity.
A schematic of mGC ligand-induced activity, phosphorylation, and destabilization, leading to the formation of the mGC–Hsp90–Cdc37 complex structurally characterized in this work. This core regulatory complex would then lead to refolding of the pseudokinase domain (PK) and reactivation of the receptor (i), recruitment of PP5 and dephosphorylation of the receptor (ii), or recruitment of E3 ligases and removal of the receptor (iii). An mGC is depicted in red, ligand in yellow, heat shock protein 90 (Hsp90) in blue and teal, Cdc37 in purple, a phosphatase in green, and an E3 ligase in orange.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Cricetulus griseus) | Chinese hamster ovary kidney cells | GIBCO | ExpiCHO | |
| Recombinant DNA reagent | pD649-GCN4-TM-GC-C_ICD (plasmid) | This paper | See: Methods - Cloning and protein expression | |
| Software, algorithm | Data collection software | SerialEM | SerialEM | |
| Software, algorithm | Data processing software | Structura Biotechnology Inc. | cryoSPARC | |
| Software, algorithm | Data sharpening software | Sanchez-Garcia et al., 2021 | DeepEMhancer | |
| Software, algorithm | Initial modeling software | Jumper et al., 2021 | AlphaFold | |
| Software, algorithm | Graphics software | Pettersen et al., 2021 | UCSF ChimeraX | |
| Software, algorithm | Modeling and refinement software | Adams et al., 2010 | Phenix | |
| Software, algorithm | Modeling and refinement software | Emsley and Cowtan, 2004 | Coot | |
| Software, algorithm | Model validation software | Chen et al., 2010 | MolProbity |
Cryo-EM data collection, refinement, and validation statistics.
| GC-C–Hsp90–Cdc37 complexPDB 8FX4EMD-29523 | GC-C–Hsp90–Cdc37 complex with DD density | |
|---|---|---|
| Data collection and processing | ||
| Nominal magnification | 45,000 | |
| Acceleration voltage (kV) | 200 | |
| Electron exposure (e-/Å2) | 58.8 | |
| Defocus range (µm) | 0.8–2.0 | |
| Pixel size (Å) | 0.9273 | |
| Symmetry imposed | C1 | |
| Final particle images | 165,635 | 48,283 |
| Map resolution FSC threshold | 0.143 | |
| Map resolution (Å) | 3.9 | 6.3 |
| Refinement | ||
| Initial model used (PDB) | 5FWK, 7ZR5, AlphaFold | |
| Model resolution FSC threshold (Å) | 0.5 | |
| Model resolution (Å) | 4.2 | |
| Model Composition | ||
| Non-hydrogen atoms | 13,478 | |
| Protein residues | 1,654 | |
| Ligands | 2 | |
| B-factors (Å2) | ||
| Protein | 119.49 | |
| Ligand | 102.85 | |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.004 | |
| Bond angles (°) | 0.914 | |
| Validation | ||
| MolProbity score | 2.14 | |
| Clashscore | 13.88 | |
| Rotamer outliers (%) | 0.67 | |
| Ramachandran plot | ||
| Favored (%) | 92.0 | |
| Allowed (%) | 7.6 | |
| Outliers (%) | 0.4 | |
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/86784/elife-86784-mdarchecklist1-v1.pdf
-
Supplementary file 1
Plasmids used in this study.
- https://cdn.elifesciences.org/articles/86784/elife-86784-supp1-v1.docx





