Binding of LncDACH1 to dystrophin impairs the membrane trafficking of Nav1.5 protein and increases ventricular arrhythmia susceptibility
Figures
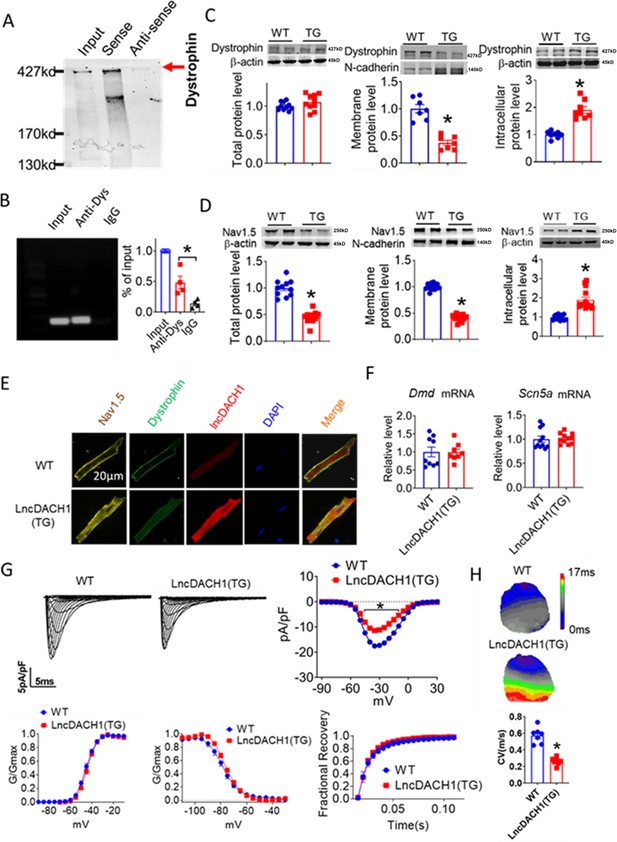
Binding of lncRNA-Dachshund homolog 1 (lncDach1) to dystrophin and the effects of cardiomyocyte-specific transgenic overexpression of lncDach1 (lncDach1-TG) on the expression and function of sodium channel.
(A) Blotting of dystrophin pulled-down by lncDach1. (B) LncDach1 precipitated by the antibody of dystrophin. N=4. *p<0.05 vs IgG by one-way ANOVA, followed by Tukey’s post-hoc analysis. (C, D) The total, membrane and intracellular levels of dystrophin and Nav1.5 by Western blot. N-cadherin is the loading control for membrane extracts. N=10–11 for total protein; N=7–14 for membrane protein; N=8–14 for intracellular protein. *p<0.05 vs wild-type (WT) group. p-values were determined by unpaired t-test. (E) Distribution of lncDach1, dystrophin, and Nav1.5 in isolated cardiomyocytes. (F) The mRNA levels of Dmd and Scn5a. N=8–11. (G) Peak INa currents, I-V curve and kinetics of INa. N=9–21 cells from three mice. *p<0.05 vs WT group. (H) Conduction velocity of perfused hearts by optical mapping recordings. N=7. *p<0.05 vs WT group. p-values were determined by unpaired t-test.
-
Figure 1—source data 1
Source data for Figure 1B-D and F-H.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig1-data1-v1.xlsx
-
Figure 1—source data 2
Uncropped and labeled gels for Figure 1A.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig1-data2-v1.pdf
-
Figure 1—source data 3
Raw unedited gels for Figure 1A.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig1-data3-v1.zip
-
Figure 1—source data 4
Uncropped and labeled gels for Figure 1C.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig1-data4-v1.pdf
-
Figure 1—source data 5
Raw unedited gels for Figure 1C.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig1-data5-v1.zip
-
Figure 1—source data 6
Uncropped and labeled gels for Figure 1D.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig1-data6-v1.pdf
-
Figure 1—source data 7
Raw unedited gels for Figure 1D.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig1-data7-v1.zip
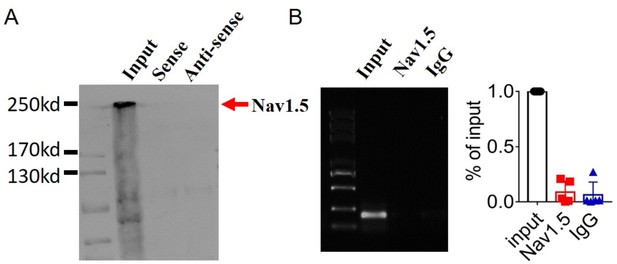
Examination of the potential interaction between lncRNA-Dachshund homolog 1 (lncDach1) and Nav1.5.
(A) Pulldown of Nav1.5 with sense and antisense sequences of lncDach1. (B) RNA immunoprecipitation of lncDach1 with anti-Nav1.5 antibody. N=4.
-
Figure 1—figure supplement 1—source data 1
Source data for Figure 1—figure supplement 1B.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig1-figsupp1-data1-v1.xlsx
-
Figure 1—figure supplement 1—source data 2
Uncropped and labeled gels for Figure 1—figure supplement 1A.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig1-figsupp1-data2-v1.pdf
-
Figure 1—figure supplement 1—source data 3
Raw unedited gels for Figure 1—figure supplement 1A.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig1-figsupp1-data3-v1.zip
-
Figure 1—figure supplement 1—source data 4
Source data for Figure 1—figure supplement 1B.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig1-figsupp1-data4-v1.xlsx
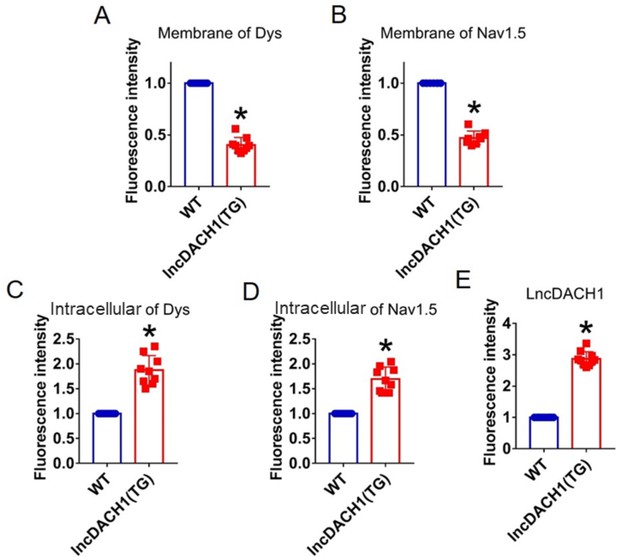
Fluorescence intensity of lncRNA-Dachshund homolog 1 (lncDach1), dystrophin, and Nav1.5 in isolated cardiomyocytes of lncDach1-TG mice.
(A, B) Membrane levels of dystrophin (dys) and Nav1.5. N=9 for dys. N=8 for Nav1.5. *p<0.05 vs wild-type (WT) group. (C, D) Intracellular levels of dystrophin and Nav1.5. N=9. *p<0.05 vs WT group. (E) Fluorescence in situ hybridization (FISH) images of LncDach1. N=10. *p<0.05 vs WT group. p-values were determined by unpaired t-test.
-
Figure 1—figure supplement 2—source data 1
Source data for Figure 1—figure supplement 2.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig1-figsupp2-data1-v1.xlsx
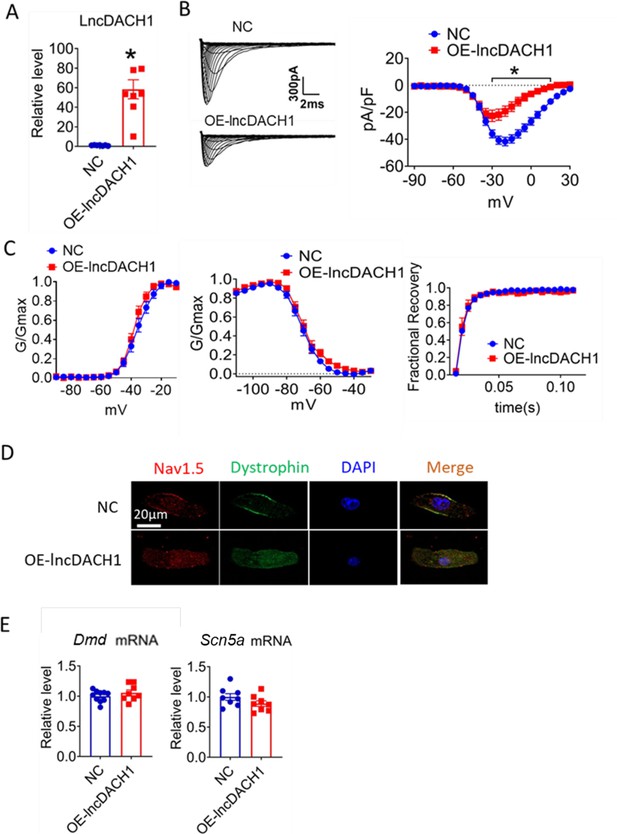
Effects of lncRNA-Dachshund homolog 1 (lncDach1) overexpression on sodium channel expression and function in cultured neonatal cardiomyocytes.
(A) Verification of the expression of lncDach1 after transfection of adenovirus carrying lncDach1. N=7–8 from three independent cultures. *p<0.05 vs NC (negative control, empty plasmid). p-values were determined by unpaired t-test. (B, C) Peak INa currents, I-V curves and kinetics of INa. N=7–25 cells from three independent cultures. *p<0.05 vs NC. (D) Distribution of Nav1.5 and dystrophin by immunofluorescent staining. (E) The mRNA levels of Dmd and Scn5a. N=8–10 from hree independent cultures.
-
Figure 2—source data 1
Source data for Figure 2A–C and E.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig2-data1-v1.xlsx
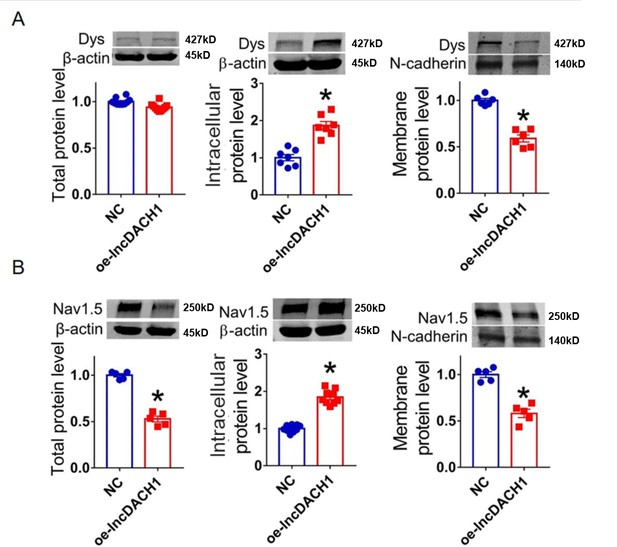
Overexpression of lncDach1(oe-lncDach1) in cultured neonatal cardiomyocytes decreased membrane dystrophin and Nav1.5 expression.
(A) The total, membrane and intracellular levels of dystrophin by Western blot. N-cadherin is the loading control for membrane extracts. N=10 for total protein; N=6 for membrane protein; N=7 for intracellular protein. *p<0.05 vs negative control (NC) group. p-values were determined by unpaired t-test. (B) The total, membrane and intracellular levels of Nav1.5 by Western blot. N-cadherin is the loading control for membrane extracts. N=5 for total protein; N=5 for membrane protein; N=9 for intracellular protein. *p<0.05 vs wild-type (WT) group. p-values were determined by unpaired t-test.
-
Figure 2—figure supplement 1—source data 1
Source data for Figure 2—figure supplement 1A–B.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig2-figsupp1-data1-v1.xlsx
-
Figure 2—figure supplement 1—source data 2
Uncropped and labeled gels for Figure 2—figure supplement 1A.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig2-figsupp1-data2-v1.pdf
-
Figure 2—figure supplement 1—source data 3
Raw unedited gels for Figure 2—figure supplement 1A.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig2-figsupp1-data3-v1.zip
-
Figure 2—figure supplement 1—source data 4
Uncropped and labeled gels for Figure 2—figure supplement 1B.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig2-figsupp1-data4-v1.pdf
-
Figure 2—figure supplement 1—source data 5
Raw unedited gels for Figure 2—figure supplement 1B.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig2-figsupp1-data5-v1.zip
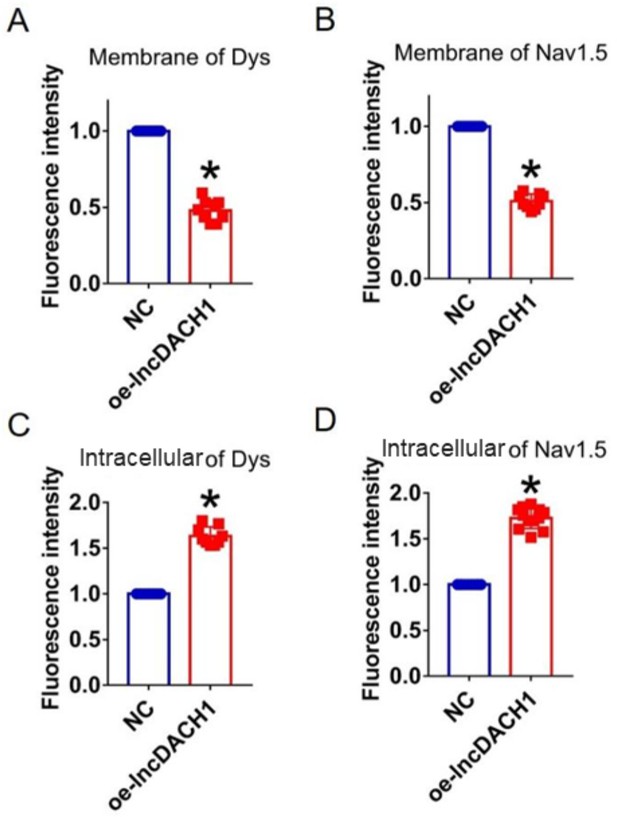
Fluorescence intensity of dystrophin and Nav1.5 in cultured neonatal cardiomyocyte overexpressing lncRNA-Dachshund homolog 1 (lncDach1).
(A, B) Membrane levels of dystrophin and Nav1.5. N=9. *p<0.05 vs negative control (NC) group. (C, D) Intracellular levels of dystrophin and Nav1.5. N=9 for dys. N=12 for Nav1.5. *p<0.05 vs NC group. P-values were determined by unpaired t-test.
-
Figure 2—figure supplement 2—source data 1
Source data for Figure 2—figure supplement 2.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig2-figsupp2-data1-v1.xlsx
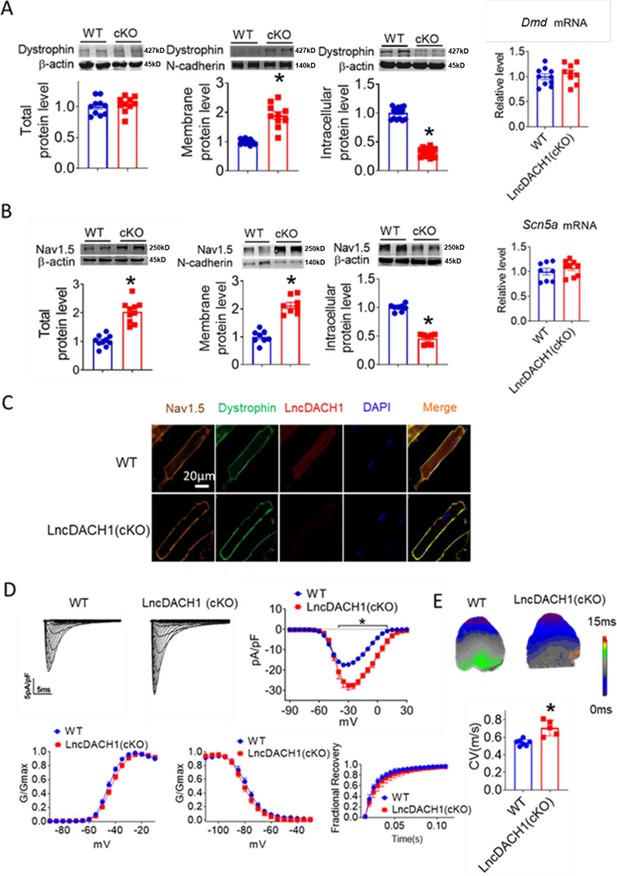
Conditional knockout of negative control (lncDach1) (lncDach1-cKO) in cardiomyocytes increased peak sodium current, membrane Nav1.5 expression.
(A) The total, membrane and intracellular levels of dystrophin by Western blot and Dmd mRNA by qRT-PCR. N-cadherin is the loading control for membrane extracts. N=10 for total protein; N=10 for membrane protein; N=15 for intracellular protein. *p<0.05 vs ild-type (WT) group. p-values were determined by unpaired t-test. (B) The total, membrane and intracellular levels of Nav1.5 by Western blot and Scn5a mRNA by qRT-PCR. N-cadherin is the loading control for membrane extracts. N=10 for total protein; N=8 for membrane protein; N=8 for intracellular protein. *p<0.05 vs WT group. p-values were determined by unpaired t-test. (C) Distribution of lncDach1, dystrophin, and Nav1.5 in isolated cardiomyocytes. (D) Peak INa currents, I-V curve, and kinetics of INa. N=7–21 cells; N=3 mice of WT; N=4 mice of LncDach1 (cKO). *p<0.05 vs WT group. (E) Conduction velocity of perfused hearts by optical mapping recordings. N=7 and 5. *p<0.05 vs WT group. p-values were determined by unpaired t-test.
-
Figure 3—source data 1
Source data for Figure 3A-B and D-E.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig3-data1-v1.xlsx
-
Figure 3—source data 2
Uncropped and labeled gels for Figure 3A.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig3-data2-v1.pdf
-
Figure 3—source data 3
Raw unedited gels for Figure 3A.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig3-data3-v1.zip
-
Figure 3—source data 4
Uncropped and labeled gels for Figure 3B.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig3-data4-v1.pdf
-
Figure 3—source data 5
Raw unedited gels for Figure 3B.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig3-data5-v1.zip
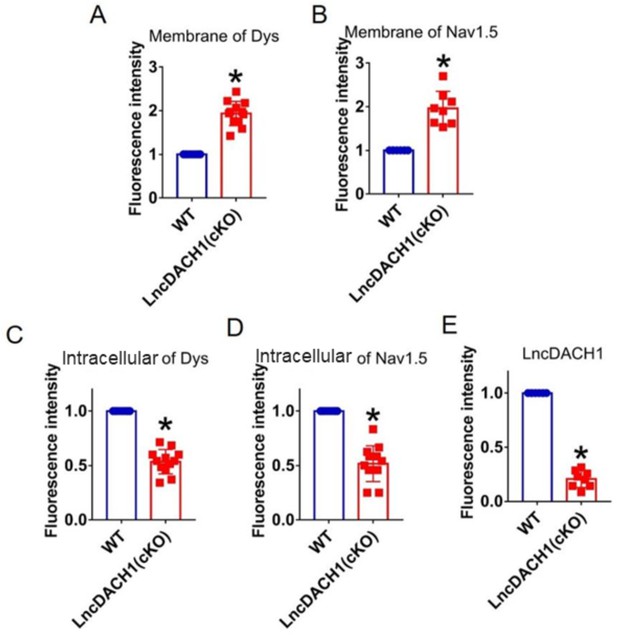
Fluorescence intensity of lncRNA-Dachshund homolog 1 (lncDach1), dystrophin, and Nav1.5 in isolated cardiomyocytes of lncDach1-cKO mice.
(A, B) Membrane levels of dystrophin (dys) and Nav1.5. N=12 for dys. N=8 for Nav1.5. *p<0.05 vs wild-type (WT) group. (C, D) Distribution of intracellular levels of dystrophin and Nav1.5. N=12. *p<0.05 vs WT group. (E) Fluorescence in situ hybridization (FISH) images of LncDach1 expression. N=8. *p<0.05 vs WT group. p-values were determined by unpaired t-test.
-
Figure 3—figure supplement 1—source data 1
Source data for Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig3-figsupp1-data1-v1.xlsx
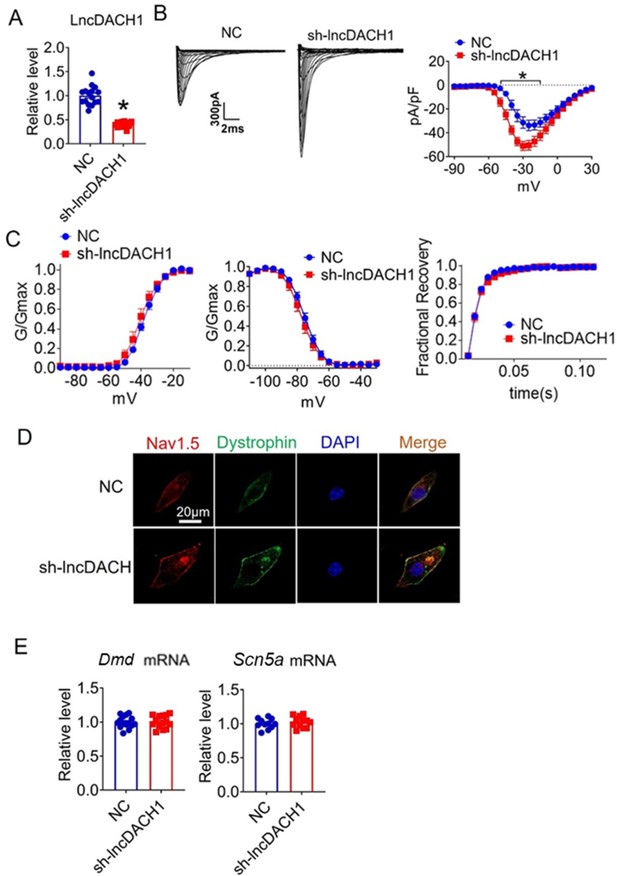
Effects of lncRNA-Dachshund homolog 1 (lncDach1) knockdown on sodium channel expression and function in cultured neonatal cardiomyocytes.
(A) Verification of the expression of lncDach1 after infection of adenovirus carrying lncDach1 shRNA. N=15 from three independent cultures. *p<0.05 vs NC (negative control, empty plasmids). p-values were determined by unpaired t-test. (B, C) Peak INa currents, I-V curve and kinetics of INa. N=8–15 from three independent cultures. *p<0.05 vs NC. (D) Distribution of Nav1.5 and dystrophin by immunofluorescent staining. (E) The mRNA levels of Dmd and Scn5a. N=11–15 from three independent cultures.
-
Figure 4—source data 1
Source data for Figure 4A–C and E.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig4-data1-v1.xlsx
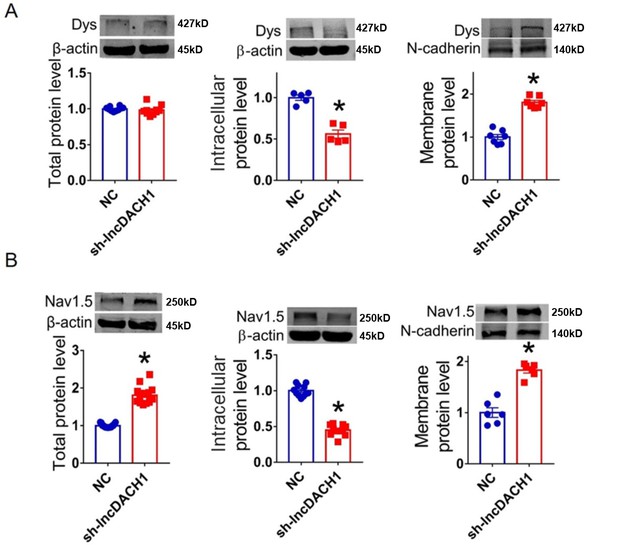
Effects of lncRNA-Dachshund homolog 1 (lncDach1) knockdown with shRNA on the protein expression of dystrophin and Nav1.5 expression in cultured neonatal cardiomyocytes.
(A) The total, membrane and intracellular levels of dystrophin by Western blot. N-cadherin is the loading control for membrane extracts. N=10 for total protein; N=7 for membrane protein; N=5 for intracellular protein. *p<0.05 vs NC group. p-values were determined by unpaired t-test. (B) The total, membrane and intracellular levels of Nav1.5 by Western blot. N-cadherin is the loading control for membrane extracts. N=15 for total protein; N=6 for membrane protein; N=11 for intracellular protein. *p<0.05 vs wild-type (WT) group. p-values were determined by unpaired t-test. Dys, dystrophin; NC, negative control; sh-lncDach1, shRNA for lncDach1.
-
Figure 4—figure supplement 1—source data 1
Source data for Figure 4—figure supplement 1A–B.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig4-figsupp1-data1-v1.xlsx
-
Figure 4—figure supplement 1—source data 2
Uncropped and labeled gels for Figure 4—figure supplement 1A.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig4-figsupp1-data2-v1.pdf
-
Figure 4—figure supplement 1—source data 3
Raw unedited gels for Figure 4—figure supplement 1A.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig4-figsupp1-data3-v1.zip
-
Figure 4—figure supplement 1—source data 4
Uncropped and labeled gels for Figure 4—figure supplement 1B.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig4-figsupp1-data4-v1.pdf
-
Figure 4—figure supplement 1—source data 5
Raw unedited gels for Figure 4—figure supplement 1B.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig4-figsupp1-data5-v1.zip
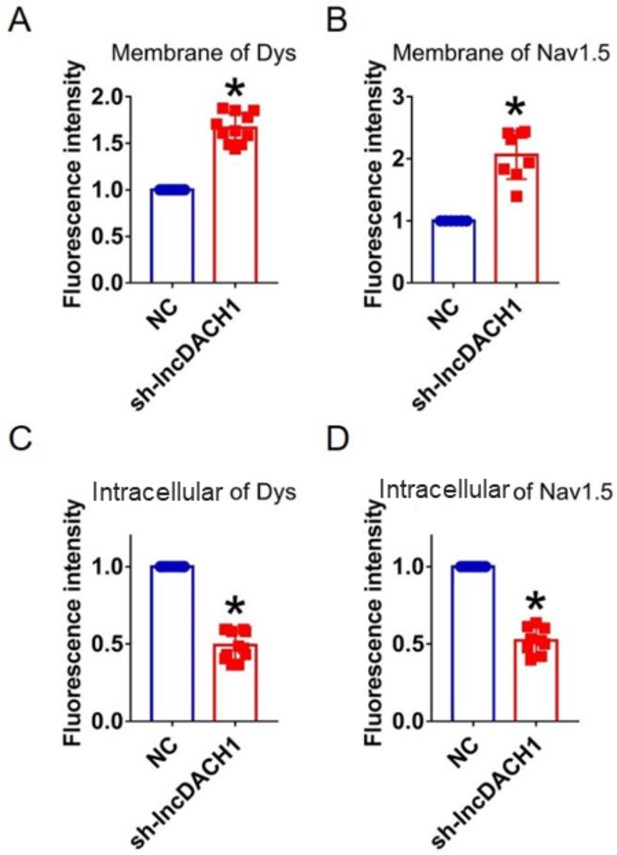
Fluorescence intensity of dystrophin and Nav1.5 in cultured neonatal cardiomyocytes after knocking down of lncRNA-Dachshund homolog 1 (lncDach1).
(A, B) Distribution of membrane levels of dystrophin and Nav1.5. N=11 for dys. N=8 for Nav1.5.*p<0.05 vs negative control (NC) group. (C, D) Distribution of intracellular levels of dystrophin and Nav1.5. N=12 for dys. N=9 for Nav1.5.*p<0.05 vs NC group. p-values were determined by unpaired t-test.
-
Figure 4—figure supplement 2—source data 1
Source data for Figure 4—figure supplement 2.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig4-figsupp2-data1-v1.xlsx
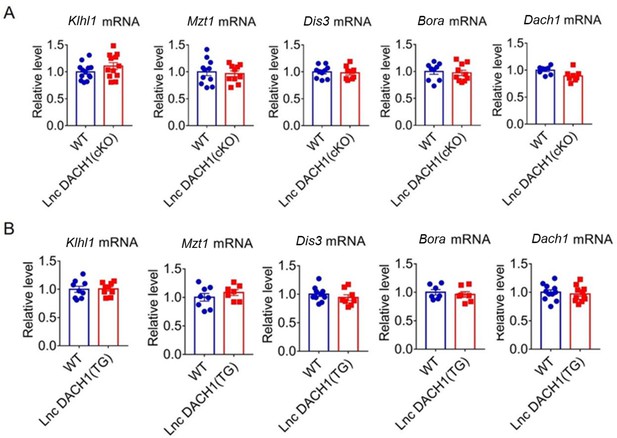
The mRNA expression of Klhl1, Bora, Mzt1, Dis3, and Dach1 in the cardiac tissue of lncRNA-Dachshund homolog 1 (lncDach1) knockout (A) and transgenic overexpression (B) mice.
N=7–12. *p<0.05 vs wild-type (WT). Data are expressed as mean ± SEM. Wt, wild-type litter mates; lncDach1(Tg), lncDach1 cardiac transgenic overexpression; lncDach1(CKO), lncDach1 cardiomyocyte conditional knockout.
-
Figure 4—figure supplement 3—source data 1
Source data for Figure 4—figure supplement 3.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig4-figsupp3-data1-v1.xlsx
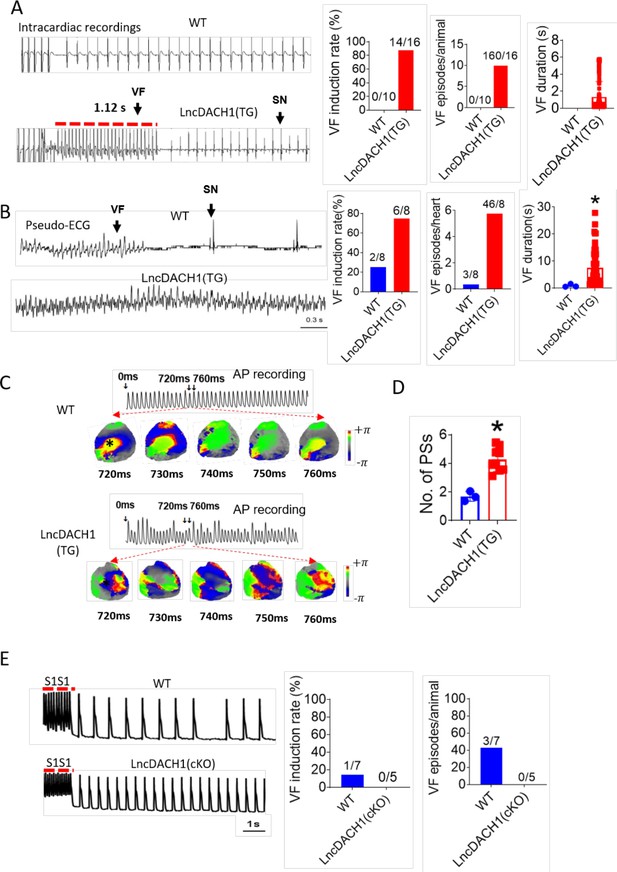
Increased arrhythmia susceptibility in lncRNA-Dachshund homolog 1 (lncDach1)-TG mice.
(A) Ventricular fibrillation (VF) induced by S1S2 pacing in intact mice. The red lines in the ECG traces indicate VF duration. N=10–16. SN: sinus rhythm (B) VF induced by S1S1 pacing of perfused hearts. (C) Break points during VT of WT and lncDach1-TG mice by optical mapping. Consecutive phase maps sampled at 10 ms interval during VF from WT and TG mice. Phase singularities (wavebreaks) are indicated by phase maps. Upper panels showed a corresponding optical recording of VF at asterisk site. N=8. (D) The number of phase singularities and dominant frequency of WT and TG mice. (E) VF induced by S1S1 pacing in perfused hearts. N=5–7 mice.
-
Figure 5—source data 1
Source data for Figure 5A-B and D-E.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig5-data1-v1.xlsx
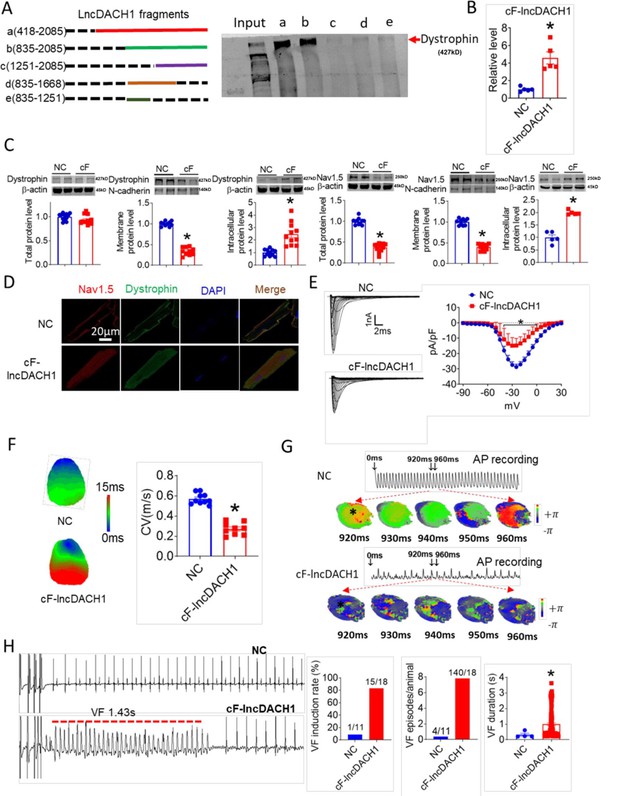
The conserved fragment of lncRNA-Dachshund homolog 1 (lncDach1) (cF-lncDach1) inhibited sodium channel function in mice.
(A) Pulldown of dystrophin by fragments of lncDach1 as indicated. (B) Verification of the expression of cF-lncDach1 after injection of adeno-virus carrying cF-lncDach1. N=5. *p<0.05 vs NC (negative control, Adeno-virus carrying empty plasmid). P-values were determined by unpaired t-test. (C) The total, membrane and intracellular levels of dystrophin and Nav1.5 by Western blot. N-cadherin is the loading control for membrane extracts. N=11–12 for total protein; N=10 for membrane protein; N=5–11 for intracellular protein. *p<0.05 vs NC group. P-values were determined by unpaired t-test. (D) Distribution of dystrophin and Nav1.5 in isolated cardiomyocytes. (E) Representative traces and I-V curve of peak INa currents. N=20–29 cells; N=3 mice of NC; N=4 mice of cF-lncDach1. *p<0.05 vs NC group. (F) Conduction velocity of perfused hearts by optical mapping recordings. N=9–10. *p<0.05 vs NC group. p-values were determined by unpaired t-test. (G) Break points during ventricular tachycardia (VT) by optical mapping. (H) Ventricular fibrillation (VF) induced by S1S2 pacing in intact mice. The induction rate, average episodes and duration of VF were determined from ECG recordings. The red lines in the ECG traces indicate VF duration. N=11 for NC; N=18 for cF-lncDach1. *p<0.05 vs NC group.
-
Figure 6—source data 1
Source data for Figure 6B–C, E–F and H.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig6-data1-v1.xlsx
-
Figure 6—source data 2
Uncropped and labeled gels for Figure 6A.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig6-data2-v1.pdf
-
Figure 6—source data 3
Raw unedited gels for Figure 6A.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig6-data3-v1.zip
-
Figure 6—source data 4
Uncropped and labeled gels for Figure 6C.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig6-data4-v1.pdf
-
Figure 6—source data 5
Raw unedited gels for Figure 6C.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig6-data5-v1.zip
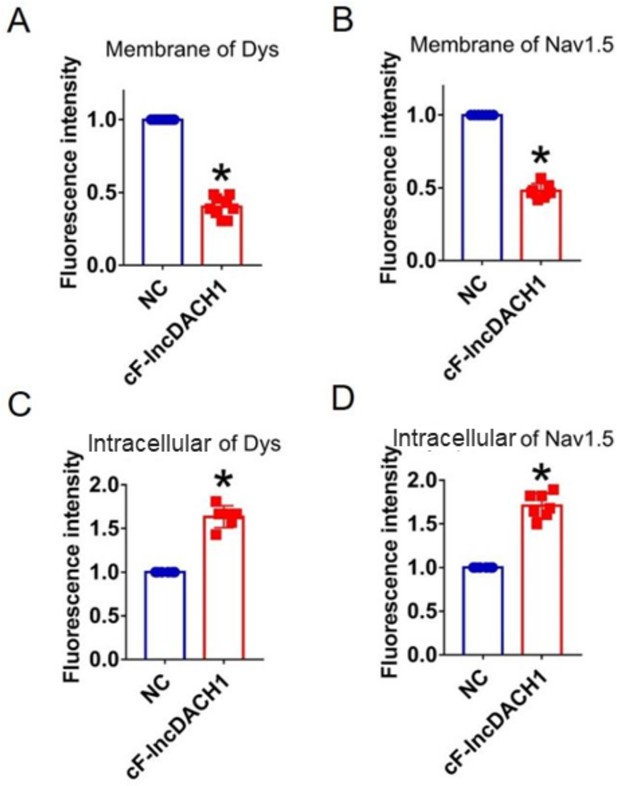
Fluorescence intensity of dystrophin and Nav1.5 in isolated cardiomyocytes overexpressing cF-lncDach1.
(A, B) Membrane levels of dystrophin (dys) and Nav1.5. N=9 for dys. N=7 for Nav1.5. *p<0.05 vs negative control (NC) group. (C, D) Intracellular levels of dystrophin and Nav1.5. N=6 for dys. N=7 for Nav1.5. *p<0.05 vs NC group. p-values were determined by unpaired t-test.
-
Figure 6—figure supplement 1—source data 1
Source data for Figure 6—figure supplement 1.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig6-figsupp1-data1-v1.xlsx
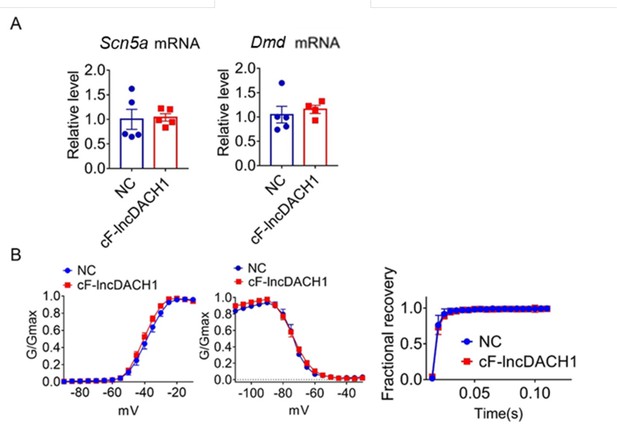
The conserved fragment of lncDach1(cF-lncDach1) inhibited sodium channel function in mice.
(A) The mRNA levels of Scn5a and Dmd. N=4–5. (B) Kinetics of INa. N=6–29 cells; N=3 mice of NC; N=4 mice of cF-lncDach1.
-
Figure 6—figure supplement 2—source data 1
Source data for Figure 6—figure supplement 2.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig6-figsupp2-data1-v1.xlsx
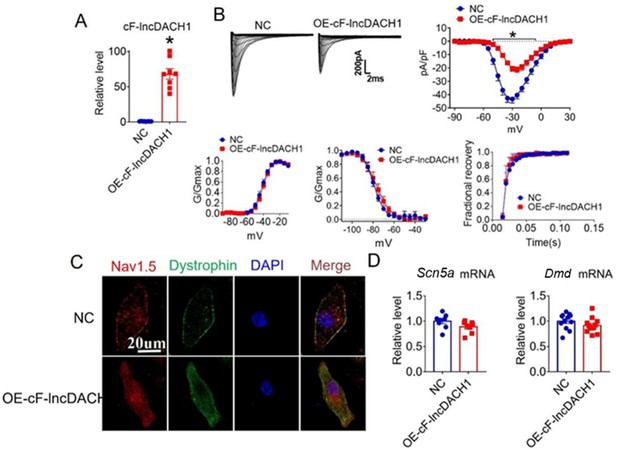
Effects of cF-lncDach1 overexpression on sodium channel expression and function in cultured neonatal cardiomyocytes.
(A) Verification of the expression of cF-lncDach1 after transfection of adenovirus carrying cF-lncDach1. N=8 from three independent cultures. *p<0.05 vs NC (negative control, empty plasmid). p-values were determined by unpaired t-test. (B) Peak INa currents, I-V curves and kinetics of INa. N=8–15 cells from three independent cultures. *p<0.05 vs NC, p-values were determined by unpaired t-test. (C) Distribution of Nav1.5 and dystrophin by immunofluorescent staining. (D) The mRNA levels of Dmd and Scn5a. N=8–12 from three independent cultures.
-
Figure 6—figure supplement 3—source data 1
Source data for Figure 6—figure supplement 3A–B and D.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig6-figsupp3-data1-v1.xlsx
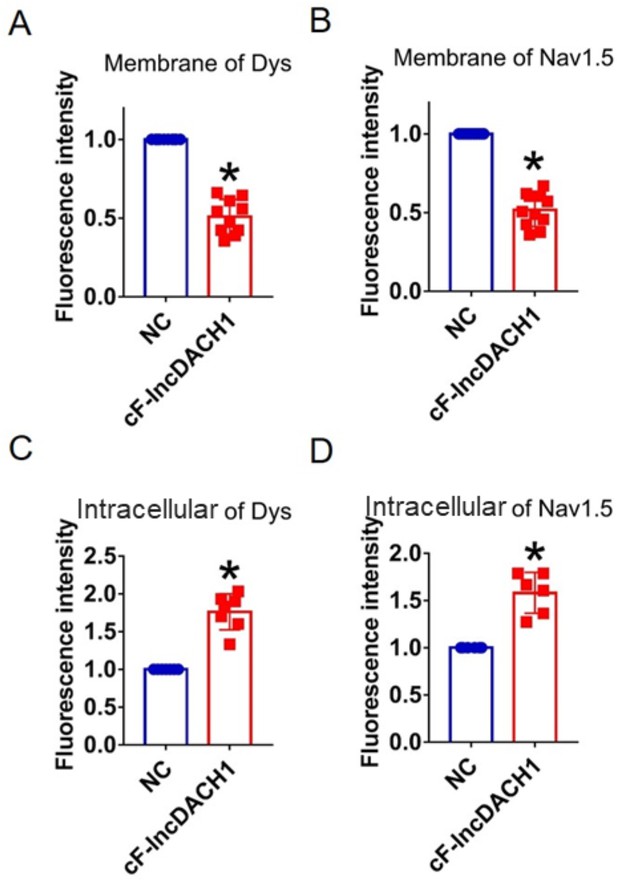
Fluorescence intensity of dystrophin and Nav1.5 in cultured neonatal cardiomyocytes overexpressing cF-lncDach1.
(A, B) Membrane levels of dystrophin and Nav1.5. N=10 for dys. N=11 for Nav1.5. *p<0.05 vs negative control (NC) group. (C, D) Intracellular levels of dystrophin and Nav1.5. N=7 for dys. N=6 for Nav1.5.*p<0.05 v s NC group. p-values were determined by unpaired t-test.
-
Figure 6—figure supplement 4—source data 1
Source data for Figure 6—figure supplement 4.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig6-figsupp4-data1-v1.xlsx
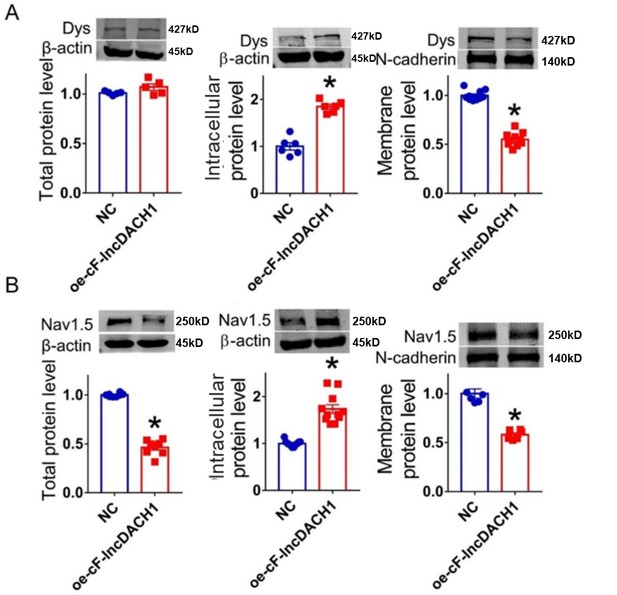
Overexpression of cF-lncDach1(oe-cF-lncDach1) in cultured neonatal cardiomyocytes decreased membrane dystrophin and Nav1.5 expression.
(A) The total, membrane and intracellular levels of dystrophin by Western blot. N-cadherin is the loading control for membrane extracts. N=5 for total protein; N=8 for membrane protein; N=6 for intracellular protein. *p<0.05 vs negative control (NC) group. p-values were determined by unpaired t-test. (B) The total, membrane and intracellular levels of Nav1.5 by Western blot. N-cadherin is the loading control for membrane extracts. N=8 for total protein; N=6 for membrane protein; N=11 for intracellular protein. *p<0.05 vs wild-type (WT) group. p-values were determined by unpaired t-test.
-
Figure 6—figure supplement 5—source data 1
Source data for Figure 6—figure supplement 5A–B.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig6-figsupp5-data1-v1.xlsx
-
Figure 6—figure supplement 5—source data 2
Uncropped and labeled gels for Figure 6—figure supplement 5A.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig6-figsupp5-data2-v1.pdf
-
Figure 6—figure supplement 5—source data 3
Raw unedited gels for Figure 6—figure supplement 5A.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig6-figsupp5-data3-v1.zip
-
Figure 6—figure supplement 5—source data 4
Uncropped and labeled gels for Figure 6—figure supplement 5B.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig6-figsupp5-data4-v1.pdf
-
Figure 6—figure supplement 5—source data 5
Raw unedited gels for Figure 6—figure supplement 5B.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig6-figsupp5-data5-v1.zip
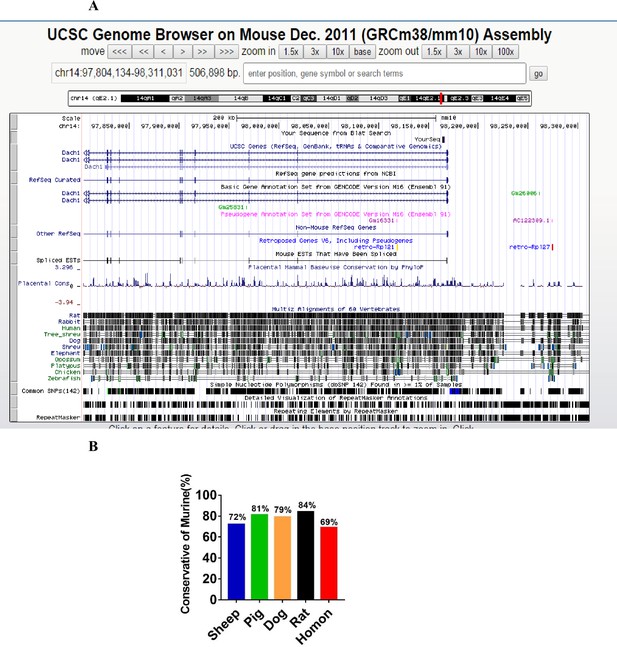
Conservation of lncDACH1.
(A) UCSC snapshot of lncRNA-Dachshund homolog 1 (lncDACH1) (B) Conservation of lncDACH1 between mice and other species.
-
Figure 6—figure supplement 6—source data 1
Source data for Figure 6—figure supplement 6B.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig6-figsupp6-data1-v1.xlsx
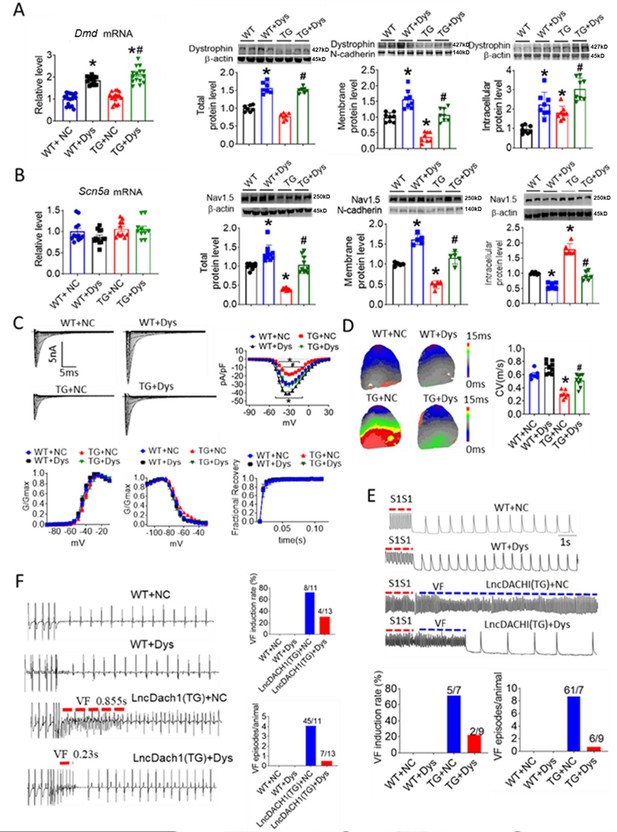
Activation of dystrophin transcription by adeno-associated virus 9 (AAV9) virus carrying dCas9-SAM system (AAV9-Dys-Act) rescued the remodeling of sodium channel in lncDach1-TG mice.
(A) The mRNA level of Dmd by real-time PCR (N=12–17) and the total, membrane and intracellular protein levels of dystrophin by western blot. N-cadherin is the loading control for membrane extracts. N=7 for total protein; N=8 for membrane protein; N=8 for intracellular protein. *p<0.05 vs egative control (NC) group. #p<0.05 vs TG group. NC, AAV9 virus carrying dCas9-SAM system with control sgRNA; Dys, AAV9 virus carrying dCas9-SAM system with sgRNA targeting dystrophin promoter. (B) The mRNA level of Scn5a by real-time PCR (N=10–12) and the total, membrane and intracellular protein levels of Nav1.5 by western blot. N-cadherin is the loading control for membrane extracts. N=10 for total protein; N=5 for membrane protein; N=6 for intracellular protein. *p<0.05 vs NC group. #p<0.05 vs TG group. (C) Representative traces, I-V curves and kinetics of peak INa currents. N=7–20 cells; N=3 mice of WT + NC; N=3 mice of WT + Dys; N=3 mice of TG + NC; N=4 mice of TG + Dys. *p<0.05 vs NC group. #p<0.05 vs TG group. (D) Conduction velocity of perfused hearts by optical mapping recordings. N=6–9. *p<0.05 vs NC group. #p<0.05 vs TG group. p-values were determined by unpaired t-test. (E) Ventricular fibrillation (VF) induced by S1S1 pacing in perfused hearts. N=7–10. (F) VF induced by S1S2 pacing in intact mice. N=10–13. The data are analyzed by one-way ANOVA followed by Tukey’s post-hoc analysis.
-
Figure 7—source data 1
Source data for Figure 7.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig7-data1-v1.xlsx
-
Figure 7—source data 2
Uncropped and labeled gels for Figure 7A.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig7-data2-v1.pdf
-
Figure 7—source data 3
Raw unedited gels for Figure 7A.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig7-data3-v1.zip
-
Figure 7—source data 4
Uncropped and labeled gels for Figure 7B.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig7-data4-v1.pdf
-
Figure 7—source data 5
Raw unedited gels for Figure 7B.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig7-data5-v1.zip
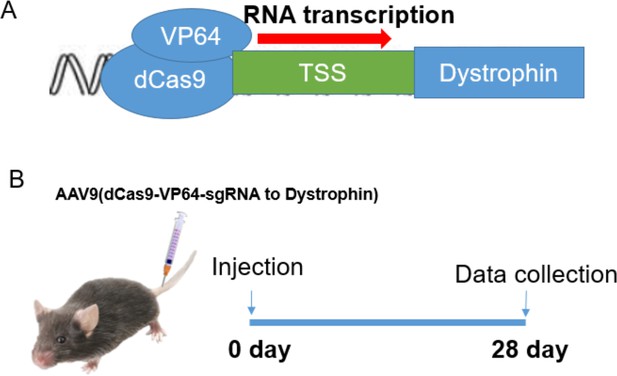
Schematic model for the construction of adeno-associated virus 9 (AAV9) virus carrying the dCas9-SAM system (A) to activate the transcription of dystrophin and tail-vein injection to mice (B).
VP64, VP64 transactivator; dCas9, deactivated CRISPR associated protein 9 nuclease. TSS, transcriptional start point.
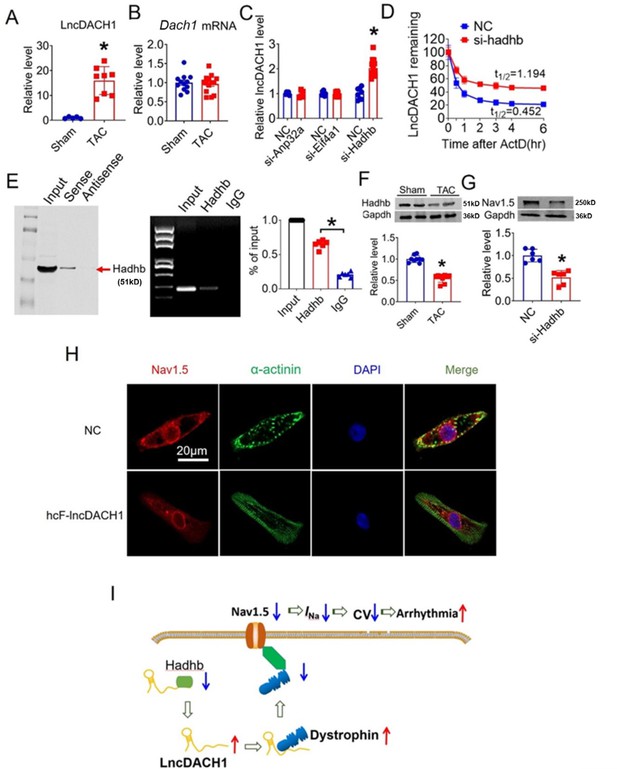
Hadhb binds to lncRNA-Dachshund homolog 1 (lncDach1) and promotes its decay.
(A) The expression level of lncDach1 in the hearts of transaortic constriction (TAC) mice. N=5–8. *p<0.05 by unpaired t-test. (B) The mRNA level of Dach1 in the hearts of TAC mice. N=12–14. (C) Effects of siRNAs for Anp32a, Eif4a1, and Hadhb on the expression of lncDach1. N=5–12 from three independent cultures. * p<0.05 by unpaired t-test. (D) The effects of Hadhb siRNA on the decay of lncDach1. N=6–15 from three independent cultures. (E) Blotting of hadhb pulled-down by lncDach1, and precipitation of lncDach1 by anti-hadhb antibody. N=4. *p<0.05 vs IgG by one-way ANOVA followed by Tukey’s post-hoc analysis. (F) The effects of heart failure on the protein expression of hadhb. N=9. *p<0.05 vs sham group. p-values were determined by unpaired t-test. (G) The effects of Hadhb siRNA on the protein expression of Nav1.5, N=6. *p<0.05 vs negative control (NC) group. p-values were determined by unpaired t-test. (H) LncDACH1 inhibits Nav1.5 in human induced pluripotent stem cells (iPS) differentiated cardiomyocytes. (I) Schematic summary of the signaling pathway of lncDACH1 and arrhythmia.
-
Figure 8—source data 1
Source data for Figure 8A–G.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig8-data1-v1.xlsx
-
Figure 8—source data 2
Uncropped and labeled gels for Figure 8E.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig8-data2-v1.pdf
-
Figure 8—source data 3
Raw unedited gels for Figure 8E.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig8-data3-v1.zip
-
Figure 8—source data 4
Uncropped and labeled gels for Figure 8F.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig8-data4-v1.pdf
-
Figure 8—source data 5
Raw unedited gels for Figure 8F.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig8-data5-v1.zip
-
Figure 8—source data 6
Uncropped and labeled gels for Figure 8G.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig8-data6-v1.pdf
-
Figure 8—source data 7
Raw unedited gels for Figure 8G.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig8-data7-v1.zip
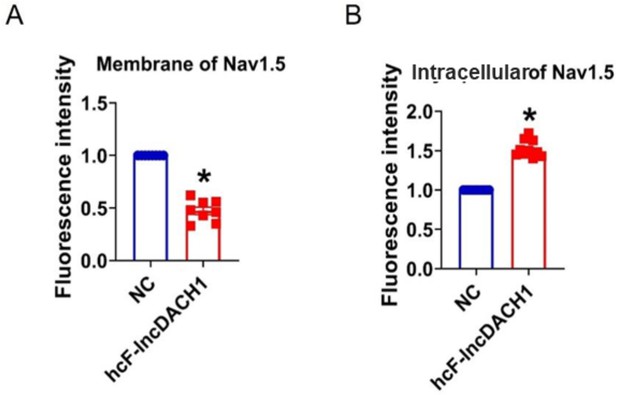
Fluorescence intensity of Nav1.5 in human induced pluripotent stem cells (iPS) differentiated cardiomyocytes overexpressing cF-lncDACH1.
(A) Membrane levels of Nav1.5. N=8 for Nav1.5. *p<0.05 vs negative control (NC) group. (B) Intracellular levels of Nav1.5. N=10 for Nav1.5.*p<0.05 vs NC group. p-values were determined by unpaired t-test.
-
Figure 8—figure supplement 1—source data 1
Source data for Figure 8—figure supplement 1.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig8-figsupp1-data1-v1.xlsx
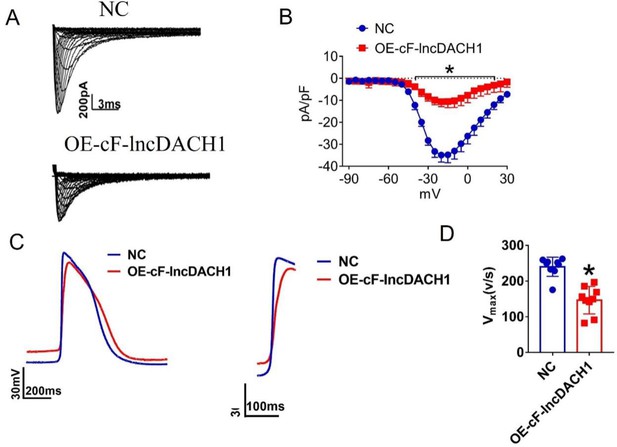
Effects of cF-lncDACH1 overexpression on sodium current and Vmax of APD in human induced pluripotent stem cells (hiPS)-CMs.
(A, B) Peak INa currents and I-V curve of INa. N=11–12 from three independent cultures. *p<0.05 vs negative control (NC). (C, D) Rising phase diagram of action potential and Vmax of ascending branch of action potential phase 0, N=9. *p<0.05 vs NC.
-
Figure 8—figure supplement 2—source data 1
Source data for Figure 8—figure supplement 2B, D.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig8-figsupp2-data1-v1.xlsx
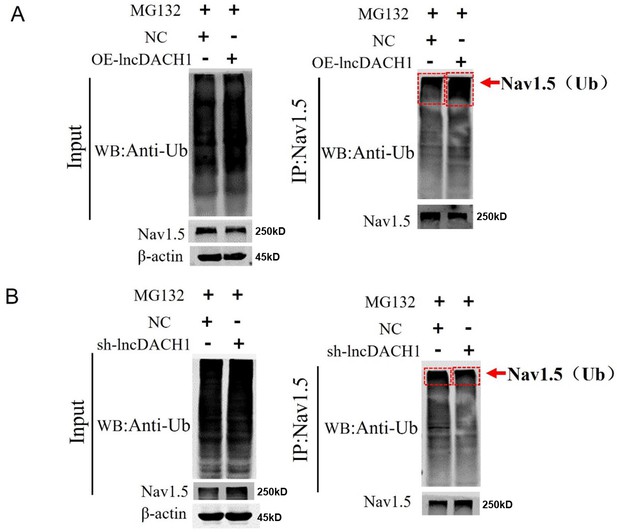
Ubiquitination of Nav1.5 in the Primary cardiomyocytes of lncRNA-Dachshund homolog 1 (lncDach1) transgenic overexpression and knockout.
(A) The ubiquitination of Nav1.5 of overexpression of lncDach1. (B) The ubiquitination of Nav1.5 of Knockdown of lncDach1.
-
Figure 8—figure supplement 3—source data 1
Uncropped and labeled gels for Figure 8—figure supplement 3A.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig8-figsupp3-data1-v1.pdf
-
Figure 8—figure supplement 3—source data 2
Raw unedited gels for Figure 8—figure supplement 3A.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig8-figsupp3-data2-v1.zip
-
Figure 8—figure supplement 3—source data 3
Uncropped and labeled gels for Figure 8—figure supplement 3B.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig8-figsupp3-data3-v1.pdf
-
Figure 8—figure supplement 3—source data 4
Raw unedited gels for Figure 8—figure supplement 3B.
- https://cdn.elifesciences.org/articles/89690/elife-89690-fig8-figsupp3-data4-v1.zip
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/89690/elife-89690-mdarchecklist1-v1.docx
-
Supplementary file 1
List of primers used in this study.
- https://cdn.elifesciences.org/articles/89690/elife-89690-supp1-v1.xlsx
-
Supplementary file 2
Alignment of murine lncDACH1.
- https://cdn.elifesciences.org/articles/89690/elife-89690-supp2-v1.docx





