Metabolic reprogramming of cancer cells by JMJD6-mediated pre-mRNA splicing associated with therapeutic response to splicing inhibitor
Figures
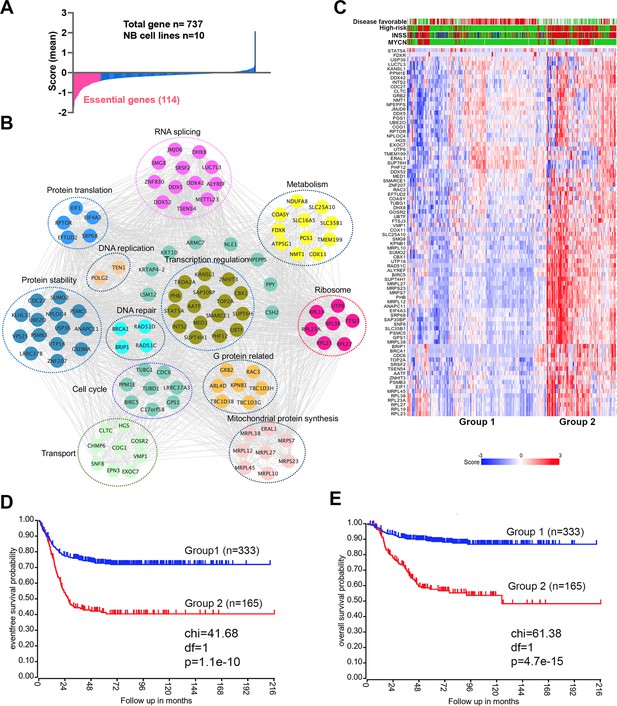
17q contains neuroblastoma dependency genes.
(A) CRISPR score for 17q genes in 10 neuroblastoma cell lines. Score <−0.4 is defined as neuroblastoma dependency genes. Data are derived from Avana sgRNA library screening (Meyers et al., 2017). (B) STRING protein interaction network showing 17q essential genes with various biological functions. (C) Heatmap by K-means clustering analysis showing 17q essential genes are highly expressed in high-risk neuroblastomas based on RNA-seq data (SEQC dataset). (D) Kaplan-Meier survival curve showing 17q essential gene signature is correlated with worse event-free survival (SEQC dataset). (E) Kaplan-Meier survival curve showing 17q essential gene signature is correlated with worse overall survival (SEQC dataset).
-
Figure 1—source data 1
17q gene list.
- https://cdn.elifesciences.org/articles/90993/elife-90993-fig1-data1-v1.xlsx
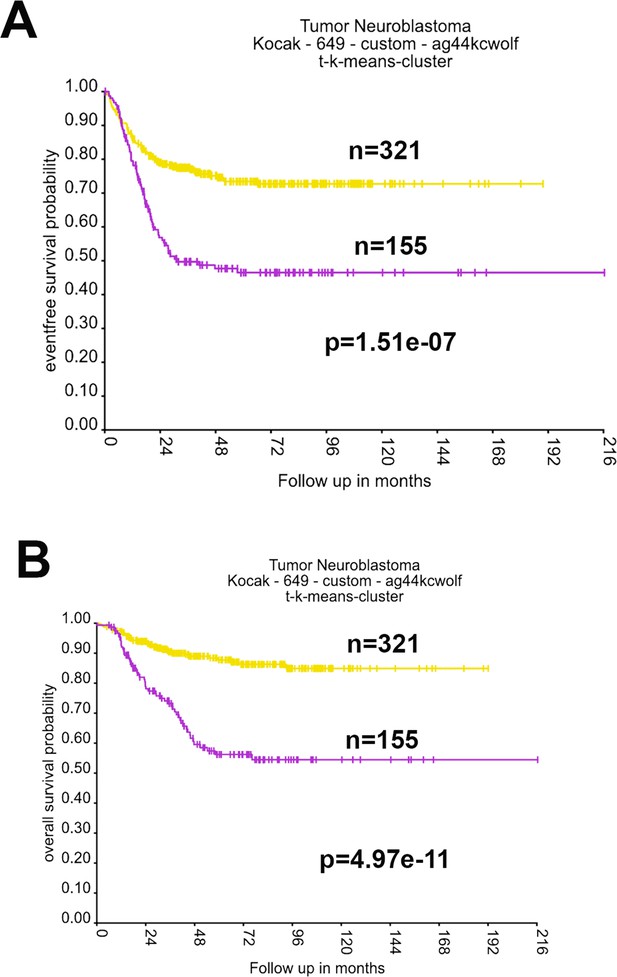
High expression of 17q essential genes is associated with worse event-free and overall survival.
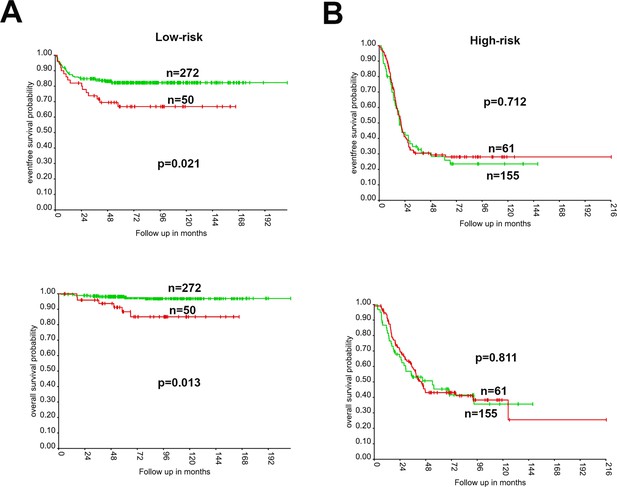
High expression of 17q essential genes is associated with worse event-free and overall survival in low-risk neuroblastomas but not high-risk neuroblastomas.
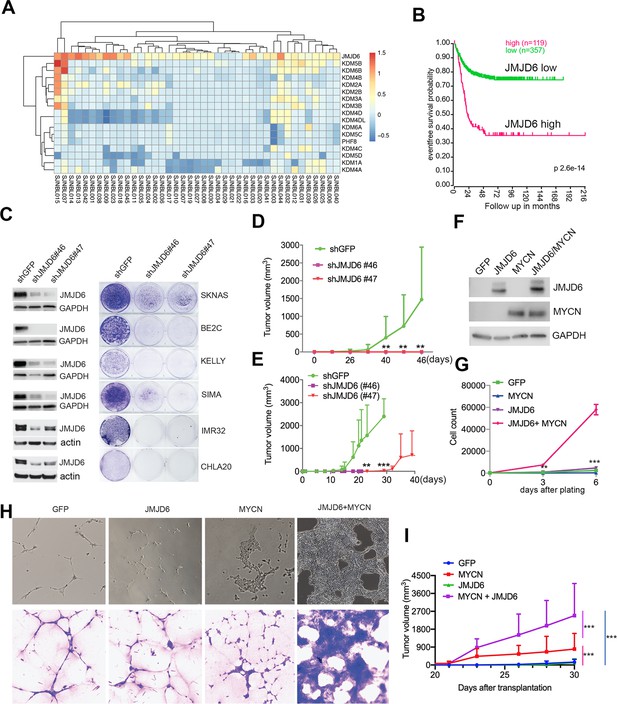
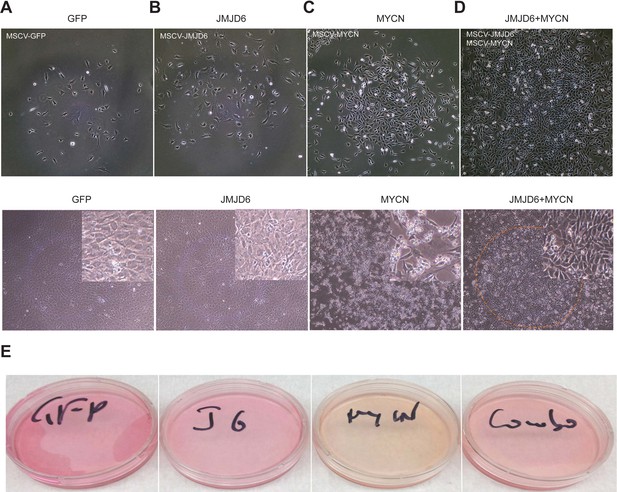
JMJD6 is required for neuroblastoma growth and facilitates MYC-mediated cellular transformation.
(A) Copy number of genes encoding JmjC domain proteins in St Jude neuroblastoma cohort (https://platform.stjude.cloud). (B) Kaplan-Meier survival curve showing high JMJD6 is correlated with worse event-free survival (SEQC RNA-seq dataset). (C) Crystal violet showing the colony staining on day 7 after JMJD6 shRNA knockdown in neuroblastoma cell lines validated by western blot (harvested at 72 hr). n=single experiment. (D) Xenograft tumor growth of BE2C (right) models with lentiviral JMJD6 shRNA knockdown. p-Value calculated by multiple unpaired t-test across each row. n=5 per group. ***p<0.001, **p<0.01. (E) Xenograft tumor growth of SK-N-AS models with lentiviral JMJD6 shRNA knockdown. p-value calculated by multiple unpaired t-test across each row. ***p<0.001, **p<0.01. (F) Western blot validating the expression of retroviral-based MYCN and JMJD6 in JoMa1 cells. (G) Cell proliferation of JoMa1 cells transduced with indicated constructs expressing GFP, JMJD6, MYCN, JMJD6+MYCN. (H) Colony formation of JoMa1 cells transduced with indicated constructs, GFP, JMJD6, MYCN, JMJD6+MYCN. Top panel showing photos taken under light microscope. Bottom panel showing cell colonies stained with crystal violet. (I) Xenograft tumor growth of JoMa1 cells transduced with indicated constructs, GFP, JMJD6, MYCN, JMJD6+MYCN. n=5 per group. p-Value calculated by multiple unpaired t-test across each row. ***p<0.001, **p<0.01. Data are Mean ± SEM.
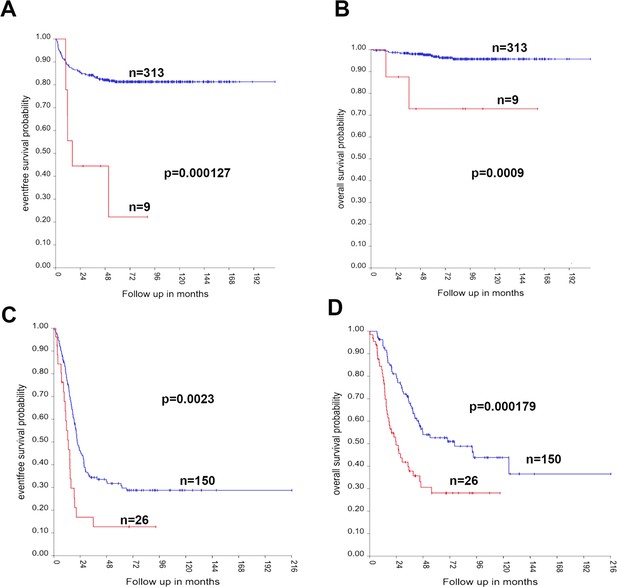
High expression of JMJD6 is associated with event-free and overall survival in both low-risk and high-risk neuroblastomas.
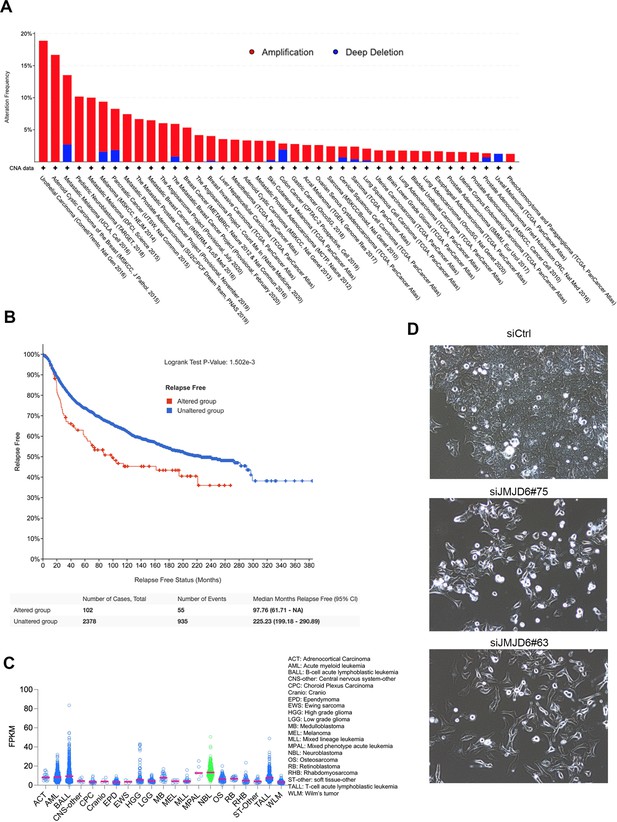
JMJD6 expression in cancers and effect on neuroblastoma differentiation.
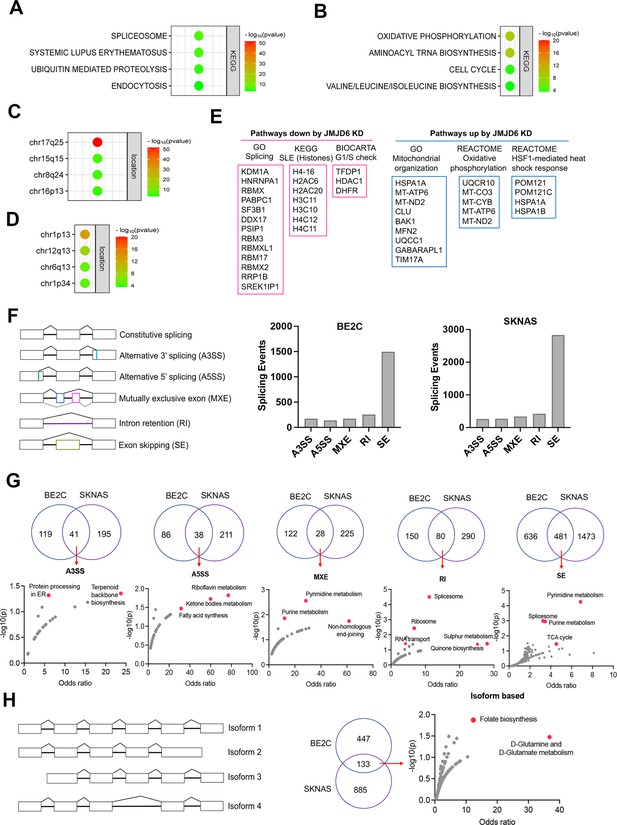
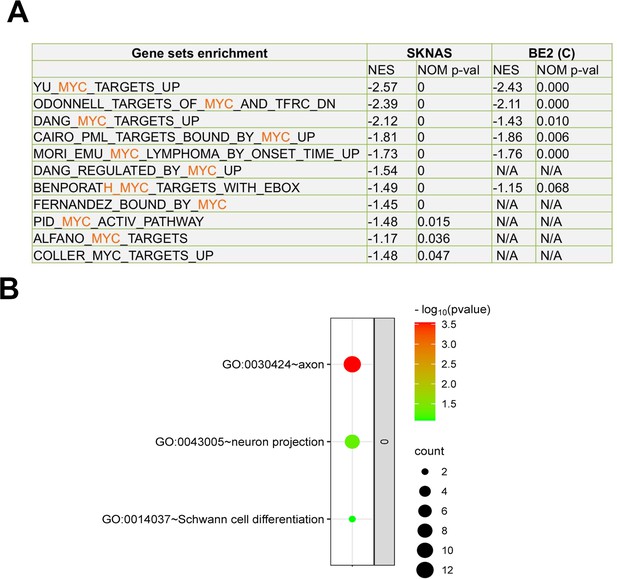
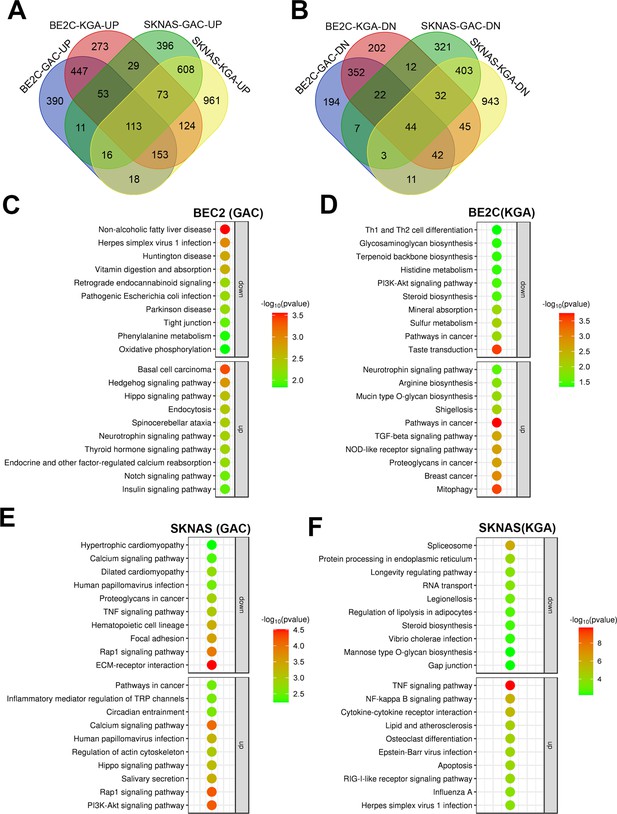
JMJD6 regulates pre-mRNA splicing of genes involved in metabolism.
(A) Pathway enrichment for JMJD6 co-dependency genes whose knockout exhibits similar phenotype with JMJD6 knockout based on re-analysis of DepMap data. (B) Pathway enrichment for genes whose knockout exhibits opposite phenotype with JMJD6 knockout based on re-analysis of DepMap data. (C) Chromosomal location enrichment for JMJD6 co-dependency genes whose knockout exhibits similar phenotype with JMJD6 knockout based on re-analysis of DepMap data. (D) Chromosomal location enrichment for genes whose knockout exhibits opposite phenotype with JMJD6 knockout based on re-analysis of DepMap data. (E) Pathway analysis for genes downregulated and upregulated (cutoff, log2FC = 1.7) by JMJD6 knockdown commonly shared in SK-NAS and BE2C cells. (F) Alternative splicing events altered by JMJD6 knockdown in BE2C and SK-N-AS cells. (G) Pathway enrichment for the genes with each splicing event commonly altered in BE2C and SK-N-AS cells after JMJD6 knockdown. (H) Isoform identification based on splicing events in BE2C and SK-N-AS cells, followed by pathway enrichment for commonly shared alterations in both cell lines.
-
Figure 3—source data 1
Differential gene expression after JMJD6 knockdown in BE2C and SKNAS cells.
- https://cdn.elifesciences.org/articles/90993/elife-90993-fig3-data1-v1.xlsx
-
Figure 3—source data 2
Pathways affected by JMJD6 knockdown in BE2C and SKNAS cells.
- https://cdn.elifesciences.org/articles/90993/elife-90993-fig3-data2-v1.xlsx
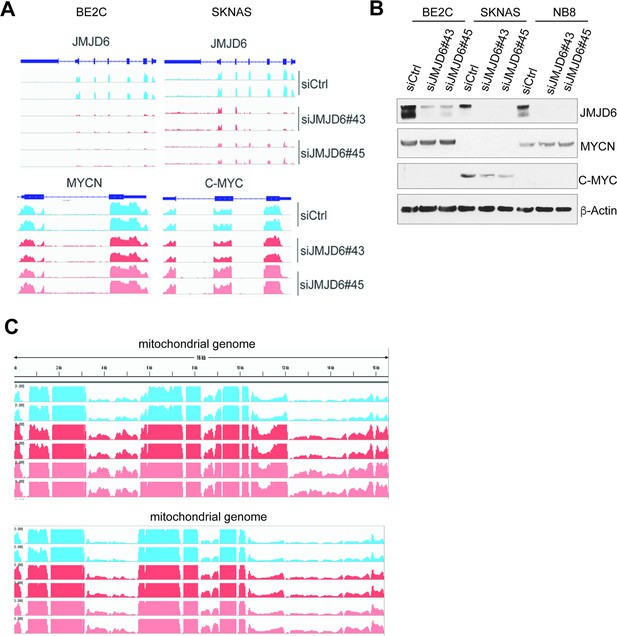
JMJD6 knockdown does not affect MYC expression but upregulates mitochondrial gene expression.
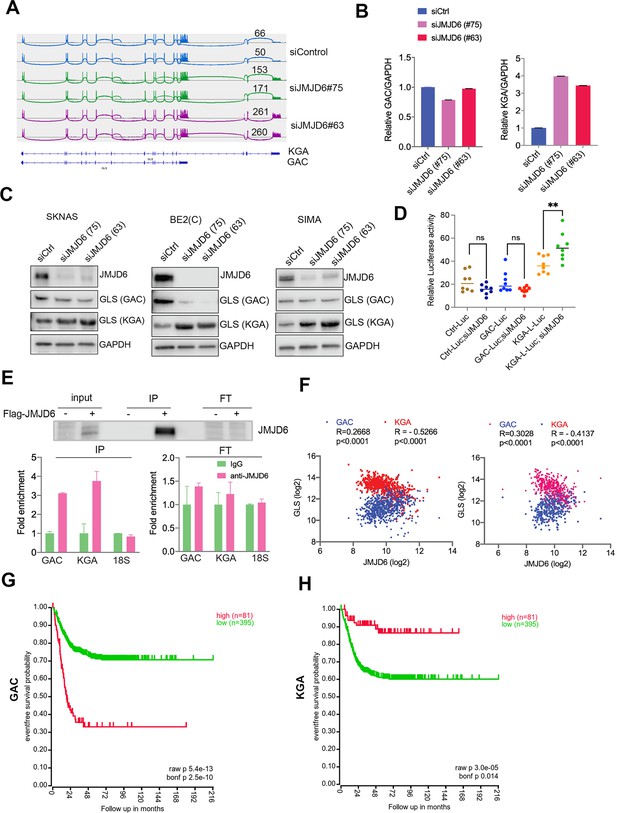
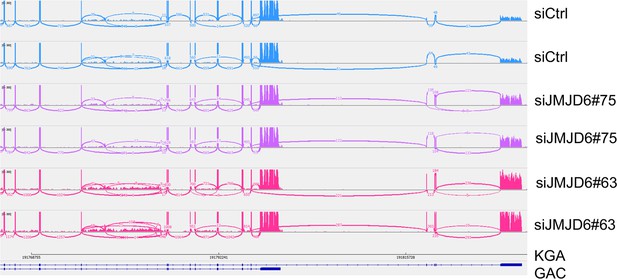
JMJD6 regulates alternative splicing of glutaminolysis gene, GLS.
(A) Sashimi plot showing the alternative splicing of GLS after JMJD6 knockdown in BE2C cells in duplicates. The number indicates the RNA-seq read counts of exon junction. (B) Real-time (RT)-polymerase chain reaction (PCR) assessing the relative expression of GAC and KGA isoforms after JMJD6 knockdown in BE2C cells in triplicates. (C) Western blot showing the expression of GAC and KGA isoforms in SK-N-AS, BE2C, SIMA after JMJD6 knockdown for 72 hr. (D) KGA- and GAC-specific reporter analysis showing only KGA-driven luciferase activity is significantly upregulated by JMJD6 knockdown. (E) RNA immunoprecipitation showing JMJD6 interaction with GLS RNA (n=single experiment). Top panel shows the western blot analysis of FLAG-tagged JMJD6 in input, immunoprecipitation (IP), and flowthrough (FT) fractions. Bottom panel shows RT-PCR (n=3) analysis of enrichment of GAC/KGA bound by JMJD6 in IP and FT fractions. (F) Spearman correlation analysis of JMJD6 and GAC/KGA expression levels in two neuroblastoma cohorts GSE45547 (left) and GSE120572 (right). (G) Kaplan-Meier curve showing the association of high or low GAC expression levels with event-free survival in a cohort of neuroblastoma (GSE45547). Expression cutoff = 3971 for GAC. (H) Kaplan-Meier curve showing the association of high or low KGA expression levels with event-free survival in a cohort of neuroblastoma (GSE45547). Expression cutoff = 7253 for KGA. Data are Mean ± SEM.
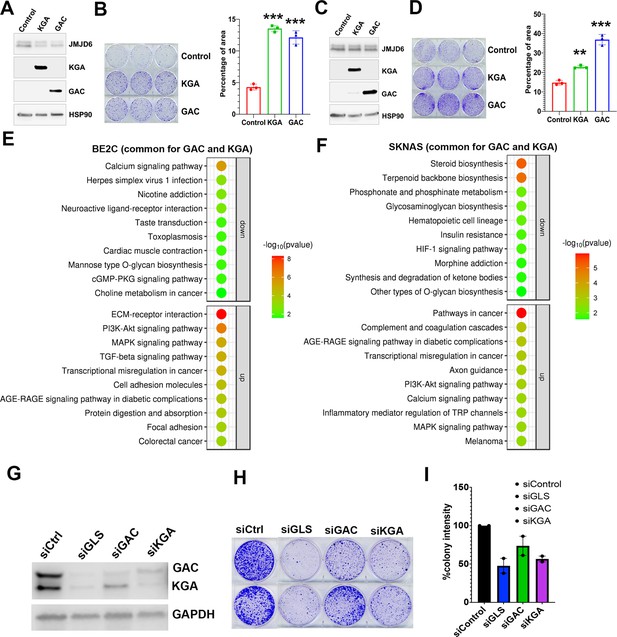
Glutaminase C (GAC) and kidney-type glutaminase (KGA) are both important for cell survival.
(A) Western blotting analysis of expression of KGA and GAC in BE2C cells with indicated antibodies. (B) Colony formation assay of BE2C cells overexpressing KGA and GAC for 7 days (left = crystal violet staining, right = quantification of cell density). n=3 per group. ***p<0.001. (C) Western blotting analysis of expression of KGA and GAC in SK-N-AS cells with indicated antibodies. (D) Colony formation assay of SK-N-AS cells overexpressing KGA and GAC for 7 days (left = crystal violet staining, right = quantification of cell density). n=3 per group. **p<0.01, ***p<0.001. (E) Bubble blot showing the pathways significantly upregulated and downregulated by both KGA and GAC in BE2C cells. (F) Bubble blot showing the pathways significantly upregulated and downregulated by both KGA and GAC in SKNAS cells. (G) Whole cell lysates (on 72 hr) subject to western blot showing the knockdown of glutaminase (GLS) (both GAC and KGA), GAC alone, and KGA alone in BE2C cells. (H) Colony-forming assay (on day 7) of BE2C cells with knockdown of GLS (both GAC and KGA), GAC alone, and KGA alone. n=2 independent experiments. (I) Quantification of colonies in (H) using ImageJ software. n=2 independent experiments. Data are Mean ± SEM.
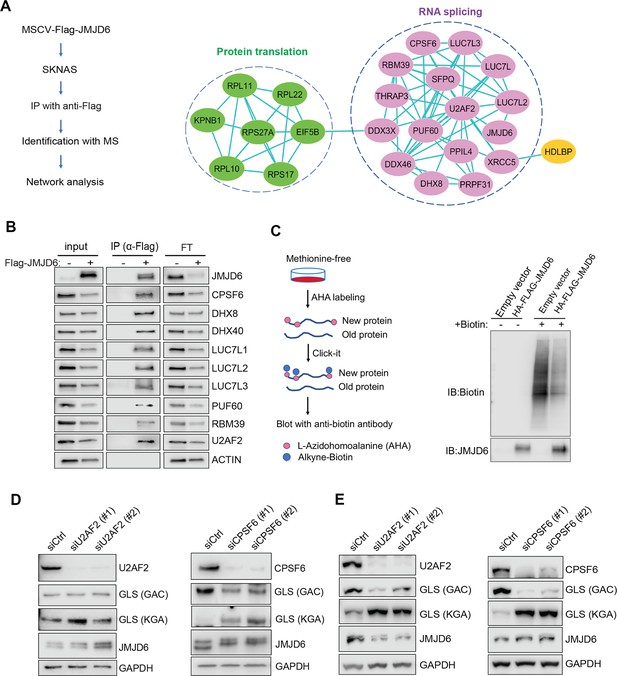
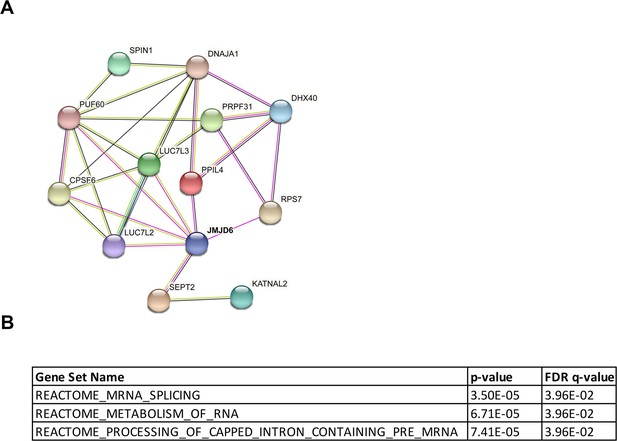
JMJD6 forms an interaction network that consists of proteins involved in splicing and protein synthesis.
(A) FLAG-tagged JMJD6 transduced into SK-N-AS cells for immunoprecipitation with anti-FLAG followed by protein identification by mass spectrometry. The interacting protein partners of JMJD6 are analyzed by STRING protein network. (B) Immunoprecipitation followed by western blot to validate the JMJD6-interacting partners in SK-N-AS cells. IP = immunoprecipitation, FT = flowthrough. n=single experiment. (C) Click-iT AHA labeling showing the newly synthesized proteins after overexpression of JMJD6 in SK-N-AS cells. n=single experiment. (D, E) Western blot showing the expression of GAC and KGA isoforms in SKNAS (D), BE2C (E), after U2AF2 and CPSF6 knockdown for 72 hr. n=single experiment.
-
Figure 6—source data 1
Mass spectrometric analysis JMJD6 interactomes in BE2C and SKNAS cells.
- https://cdn.elifesciences.org/articles/90993/elife-90993-fig6-data1-v1.xlsx
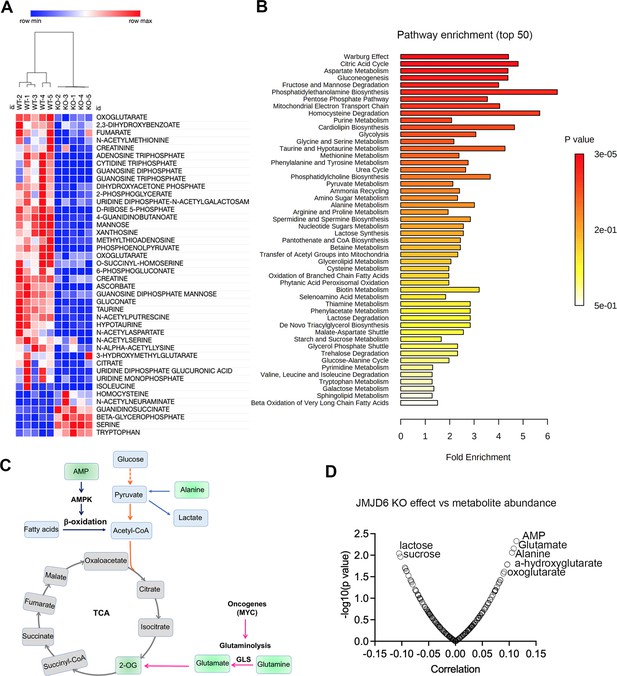
JMJD6 regulates production of citric acid cycle intermediates and NTP.
(A) Heatmap showing the metabolites differentially expressed in SK-N-AS cells (n=5) after JMJD6 knockout (n=5) based on liquid chromatography with tandem mass spectrometry (LC-MS/MS) analysis. (B) Pathway analysis of metabolites downregulated by JMJD6 knockout. (C) Pathway cartoon showing the connections of tricarboxylic acid (TCA), glycolysis, glutaminolysis, and β-oxidation. (D) Correlation of metabolite abundance with JMJD6 dependency. The positive correlation indicates that the higher the abundance of metabolites, the more resistance of cells to JMJD6 knockout. On the contrary, the negative correlation indicates the higher the abundance of metabolites, the more sensitive of cells to JMJD6 knockout.
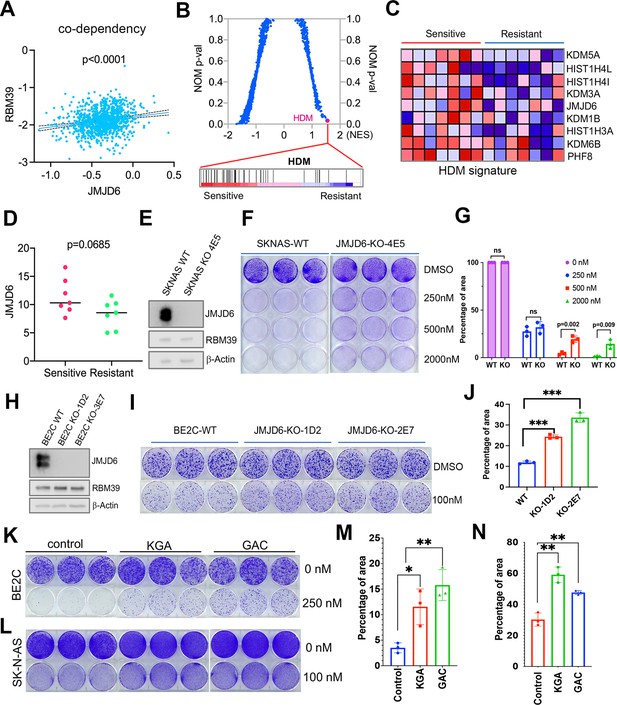
JMJD6-GAC pathway regulates the response of neuroblastoma cells to indisulam treatment.
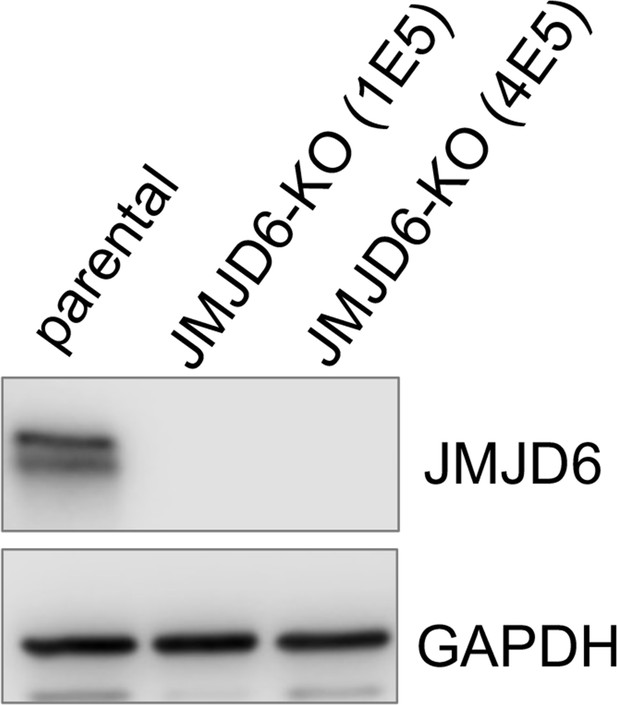
(A) Spearman correlation of effects of JMJD6 knockout and RBM39 knockout demonstrating the co-dependency of JMJD6 and RBM39 from DepMap CRISPR screening data (n=1086). Each dot represents one cell line. (B) Gene set enrichment analysis (GSEA) for indisulam sensitive vs resistant neuroblastoma cell lines based on CTD2 (Cancer Target Discovery and Development) data showing histone lysine demethylase gene signature is the one that is significantly associated with indisulam response. (C) Heatmap from GSEA (B) showing the individual genes in indisulam-sensitive and -resistant cells. (D) JMJD6 expression in indisulam-sensitive and -resistant neuroblastoma cells. p-Value calculated by Student’s t-test. (E) Western blot showing JMJD6 knockout in SK-N-AS cells using indicated antibodies. (F) Colony formation of SK-N-AS cells in triplicates with or without JMJD6 knockout treated with different concentrations of indisulam for 7 days, stained with crystal violet. n=3 per group. (G) Quantification of cell density by using ImageJ software from (F) (n=triplicates). ns = not significant. **p<0.001, ***p<0.0001. (H) Western blot showing JMJD6 knockout in BE2C cells using indicated antibodies. (I) Colony formation of BE2C cells in triplicates with or without JMJD6 knockout treated with 100 nM of indisulam for 5 days, stained with crystal violet. n=3 per group. (J) Quantification of cell density by using ImageJ software from (I) (n=triplicates). ns = not significant. **p<0.001, ***p<0.0001. (K) Colony formation of BE2C cells in triplicates with KGA and GAC overexpression treated with 250 nM of indisulam for 5 days, stained with crystal violet. n=3 per group. (L) Colony formation of SK-N-AS cells in triplicates with KGA and GAC overexpression treated with 100 nM of indisulam for 7 days, stained with crystal violet. (M) Quantification of cell density by using ImageJ software from (K) (n=triplicates). *p<0.01, **p<0.001. (N) Quantification of cell density by using ImageJ software from (L) (n=triplicates). **p<0.001. Data are Mean ± SEM.
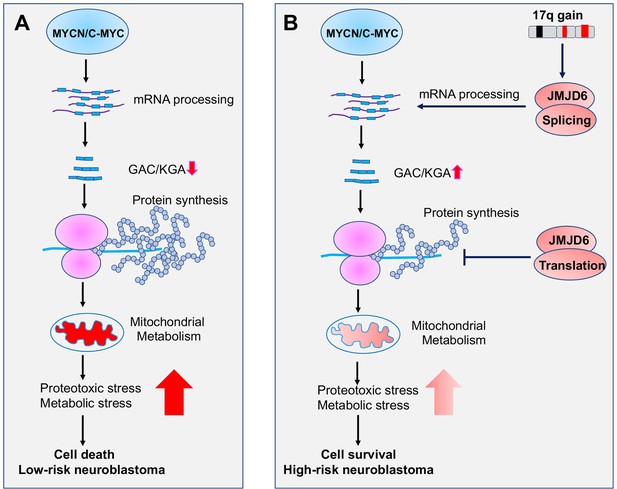
Working mechanism of JMJD6 in MYC-driven neuroblastoma.
Overactive MYC drives high-load of gene transcription, enhanced protein synthesis, and high rate of metabolism, leading to detrimental cellular stresses and consequent cell death (Model A). However, when 17q is amplified, high levels of JMJD6 and other proteins encoded by 17q genes physically interact with the splicing and translational machineries, enhancing pre-mRNA splicing of metabolic genes such as glutaminase (GLS) and inhibiting global protein synthesis, respectively, leading to reduced detrimental stresses and enhanced cancer cell survival and tumorigenesis (Model B). The high levels of JMJD6 predicts high dependency of RBM39, which are more sensitive to indisulam treatment.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Anti-GAPDH (rabbit polyclonal) | Cell Signaling Technology | 5174s,RRID:AB_10622025 | WB 1:1000 |
| Antibody | Anti-MYCN (mouse monoclonal) | Santa Cruz Biotechnology | 53993, RRID:AB_831602 | WB 1:1000 |
| Antibody | Anti-FLAG (mouse monoclonal) | Sigma | F1804, RRID:AB_262044 | WB 1:1000 |
| Antibody | Anti-Biotin (rabbit polyclonal) | Bethyl Laboratories | A150-109A, RRID:AB_67327 | WB 1:1000 |
| Antibody | Anti-ACTIN (mouse monoclonal) | Sigma | A3854, RRID:AB_262011 | WB 1:1000 |
| Antibody | Anti-PUF60 (rabbit polyclonal) | Thermo Fisher | PA5-21411, RRID:AB_11154782 | WB 1:1000 |
| Antibody | Anti-U2AF2 (rabbit polyclonal) | Novus Biologicals | NBP2-04140 | WB 1:1000 |
| Antibody | Anti-CPSF6 (rabbit polyclonal) | Bethyl Laboratories | A301-357A, RRID:AB_937783 | WB 1:1000 |
| Antibody | Anti-DHX40 (rabbit polyclonal) | Novus Biologicals | NBP1-91834, RRID:AB_11040145 | WB 1:1000 |
| Antibody | Anti-DHX8 (rabbit recombinant monoclonal) | Abcam | AB181074 | WB 1:1000 |
| Antibody | Anti-LUC7L1 (rabbit polyclonal) | Novus Biologicals | NBP2-56401 | WB 1:1000 |
| Antibody | Anti-LUC7L2 (rabbit polyclonal) | Novus Biologicals | NBP2-33621 | WB 1:1000 |
| Antibody | Anti-LUC7L3 (rabbit polyclonal) | Novus Biologicals | NBP1-88053, RRID:AB_11033957 | WB 1:1000 |
| Antibody | Anti-RBM39 (rabbit polyclonal) | ATLAS | HPA0015191, RRID:AB_1079749 | WB 1:1000 |
| Antibody | Anti-GLS (KGA-specific) (rabbit polyclonal) | Proteintech | 20170–1-AP, RRID:AB_10665373 | WB 1:1000 |
| Antibody | Anti-GLS (GAC-specific) (rabbit polyclonal) | Proteintech | 19958–1-AP, RRID:AB_10640899 | WB 1:1000 |
| Antibody | Anti-JMJD6 (rabbit polyclonal) | ATLAS | HPA059156, RRID:AB_2683934 | WB 1:1000 |
| Antibody | Anti-JMJD6 (mouse monoclonal) | Santa Cruz Biotechnology | sc-28348, RRID:AB_628185 | WB 1:1000 |
| Antibody | M2 anti-FLAG beads (mouse monoclonal) | Sigma | M8823, RRID:AB_2637089 | Antibody-conjugated beads |
| Cell line (Homo sapiens) | KELLY | ECACC | 92110411, RRID:CVCL_2092 | Neuroblastoma cell line, human, pediatric |
| Cell line (Homo sapiens) | SIMA | DSMZ | ACC164, RRID:CVCL_1695 | Neuroblastoma cell line, human, pediatric |
| Cell line (Homo sapiens) | BE2C | ATCC | CRL2268, RRID:CVCL_0529 | Neuroblastoma cell line, human, pediatric |
| Cell line (Homo sapiens) | IMR32 | ATCC | CCL127, RRID:CVCL_0346 | Neuroblastoma cell line, human, pediatric |
| Cell line (Homo sapiens) | SK-N-AS | ATCC | CRL2137, RRID:CVCL_6602 | Neuroblastoma cell line, human, pediatric |
| Cell line (Homo sapiens) | CHLA20 | COG | RRID:CVCL_6602 | Neuroblastoma cell line, human, pediatric |
| Cell line (Homo sapiens) | HEK293T/293T | ATCC | CRL3216, RRID:CVCL_0063 | Embryonic kidney, human |
| Cell line (Mus musculus) | NIH3T3 | ATCC | CRL1658, RRID:CVCL_0594 | Fibroblast cell line, mouse |
| Cell line (Mus musculus) | JoMa1 | Dr Schulte (Department of Pediatric Oncology and Hematology, University Children’s Hospital Essen, Essen, Germany) | Neural crest cell line, mouse | |
| Chemical compound, drug | Indisulam | MedKoo Biosciences | MedKoo Cat#: 201540 | RBM39 inhibitor |
| Commercial assay or kit | PowerPlex 16 HS System | Promega | DC2101 | Used for short tandem repeat (STR) profiling of all human-derived cell lines |
| Commercial assay or kit | LookOut Mycoplasma PCR Detection Kit | Sigma-Aldrich | MP0035 | Used for mycoplasma screening for all cell lines |
| Commercial assay or kit | JumpStart Taq DNA Polymerase | Sigma-Aldrich | D9307 | Used for mycoplasma screening for all cell lines |
| Commercial assay or kit | Rneasy Plus Mini Kit | QIAGEN | 74136 | For isolating RNA from cells |
| Commercial assay or kit | Superscript IV First Strand Synthesis System | Invitrogen | 1809105 | Generating cDNA from RNA |
| Commercial assay or kit | PowerUp SYBR Green Master Mix | Applied Biosystems | A25743 | Master mix for real-time PCR |
| Commercial assay or kit | Click-iT AHA (L-azidohomoalanine) | Thermo Fisher | C10102 | Kit component used for Click-iT Metabolic labeling of nascent proteins |
| Commercial assay or kit | EZQ Protein Quantification Kit | Thermo Fisher | R33200 | Kit component used for Click-iT Metabolic labeling of nascent proteins |
| Commercial assay or kit | Biotin-alkyne (PEG4 carboxamide-propargyl biotin) | Thermo Fisher | B10185 | Kit component used for Click-iT Metabolic labeling of nascent proteins |
| Commercial assay or kit | PEIpro | Polyplus | 115--010 | Transfection reagent; used at 2:1 (µL:µg of DNA) |
| Commercial assay or kit | RNAiMAX | Invitrogen | 13778100 | RNAi transfection reagent; 7 µL used per 25 µM siRNA oligo |
| Gene (Homo sapiens) | JMJD6 | NCBI | NM_001081461.2 | |
| Gene (Homo sapiens) | GLS (GAC isoform) | NCBI | NM_014905.5 | |
| Gene (Homo sapiens) | GLS (KGA isoform) | NCBI | NM_001256310.2 | |
| Gene (Mus musculus) | Mycn | NCBI | NM_001293228.2 | |
| Other | N2-Supplement | Invitrogen | 17502-048 | Supplement neural crest culture medium for JoMa1 cells |
| Other | B27-Supplement | Invitrogen | 17504-044 | Supplement neural crest culture medium for JoMa1 cells |
| Other | Chick-Embryo-Extract | Gemini Bio-Products | Supplement neural crest culture medium for JoMa1 cells | |
| Other | 4-OH-tamoxifen | Sigma | H7904 | Supplement neural crest culture medium to ensure nuclear localization of c-MycERT in JoMa1 cells |
| Other | cOmplete Protease Inhibitor | Sigma | 11836170001 | Protease inhibitor for immunoprecipitation |
| Other | PhosSTOP | Sigma | 4906845001 | Phosphatase inhibitor for immunoprecipitation |
| Other | FLAG peptide | St Jude | Used for elution of FLAG-tagged peptides during immunoprecipitation; 3 µL of stock FLAG peptide at 5 µg/µL per 100 µL of TBS buffer | |
| Other | Rnasin Rnase inhibitor | Promega | N2511 | Use at 100 U/mL for RNA immunoprecipitation |
| Other | Vanadyl ribonucleoside complexes solution | Sigma | 94742 | Use at 2 mM for RNA immunoprecipitation |
| Other | Proteinase K | Ambion | AM2548 | Digestion of protein in RNA immunoprecipitation |
| Other | Phenol-chloroform-isoamyl alcohol mixture | Sigma | 77618 | Precipitation of nucleotides in RNA immunoprecipitation |
| Other | GlycoBlue | Ambion | AM9516 | Recovery of RNA in RNA immunoprecipitation |
| Recombinant DNA reagent | MSCV-IRES-GFP (plasmid) | St Jude Vector Core | ||
| Recombinant DNA reagent | MSCV-IRES-mCherry (plasmid) | St Jude Vector Core | ||
| Recombinant DNA reagent | MSCV-JMJD6-IRES-GFP (plasmid) | This paper | ||
| Recombinant DNA reagent | MSCV-Mycn-IRES-mCherry (plasmid) | This paper | ||
| Recombinant DNA reagent | MSCV-CMV-CMV-FLAG-HA-JMJD6 (plasmid) | Addgene | 31358 | |
| Recombinant DNA reagent | pMD-old-gag-pol (plasmid) | St Jude Vector Core | ||
| Recombinant DNA reagent | VSV-G (plasmid) | St Jude Vector Core | ||
| Recombinant DNA reagent | pGenLenti (plasmid) | Genscript | Lentiviral expression vector used to express cDNA sequence of either GAC or KGA isoform of GLS gene | |
| Recombinant DNA reagent | TRC lentiviral-based shRNA knockdown plasmids to JMJD6 ‘sh#46’ | Horizon Discovery | RHS3979-201781036 | TTAAACCAGGTAATAGCTTCG |
| Recombinant DNA reagent | TRC lentiviral-based shRNA knockdown plasmids to JMJD6 ‘sh#47’ | Horizon Discovery | RHS3979-201781037 | ATCTTCACTGAGTAGCCATCG |
| Recombinant DNA reagent | shControl (pLKO.1) | St Jude Vector Core | ||
| Recombinant DNA reagent | Lentiviral helper plasmids | St Jude Vector Core | pCAG-kGP1-1R | |
| Recombinant DNA reagent | Lentiviral helper plasmids | St Jude Vector Core | pCAG4-RTR2 | |
| Recombinant DNA reagent | pMaxGFP | Lonza | For CRISPR-Cas9-mediated editing | |
| Peptide, recombinant protein | EGF | Invitrogen | Recombinant protein, media supplement | EGF |
| Peptide, recombinant protein | FGF | Invitrogen | Recombinant protein, media supplement | FGF |
| Peptide, recombinant protein | Cas9 protein | St Jude Protein Production Core | Recombinant protein, peptide | Cas9 protein |
| Sequence-based reagent | 18S rRNA F | IDT | RT-PCR primers | GCTTAATTTGACTCAACACGGGA |
| Sequence-based reagent | 18S rRNA R | IDT | RT-PCR primers | AGCTATCAATCTGTCAATCCTGTC |
| Sequence-based reagent | GLS-GACiso_F | IDT | RT-PCR primers | GAGGTGCTGGCCAAAAAGCCT |
| Sequence-based reagent | GLS-GACiso_R | IDT | RT-PCR primers | AGGCATTCGGTTGCCCAAACT |
| Sequence-based reagent | GLS-KGAiso_F | IDT | RT-PCR primers | CTGCAGAGGGTCATGTTGAA |
| Sequence-based reagent | GLS-KGAiso_R | IDT | RT-PCR primers | ATCCATGGGAGTGTTATTCCA |
| Sequence-based reagent | KGA_set2_F | IDT | RT-PCR primers | GCAGCCTCCAGGTGCTTTCA |
| Sequence-based reagent | KGA_set2_R | IDT | RT-PCR primers | GTAATGGGAGGGCAGTGGCA |
| Sequence-based reagent | KGA_set3_F | IDT | RT-PCR primers | TGCCCGACACTGCCCTTTAG |
| Sequence-based reagent | KGA_set3_R | IDT | RT-PCR primers | CCTGCCAGACAGACAACAGCA |
| Sequence-based reagent | GAC_set2_F | IDT | RT-PCR primers | TGCTTCTCAAGGCCTTACTGC |
| Sequence-based reagent | GAC_set2_R | IDT | RT-PCR primers | AGGCATTCGGTTGCCCAAACT |
| Sequence-based reagent | GAC_set3_F | IDT | RT-PCR primers | CCTTCTAGAGGTGCTGGCCAAA |
| Sequence-based reagent | GAC_set3_R | IDT | RT-PCR primers | TGCAACACAAATATGCAGTAAGGC |
| Sequence-based reagent | siRNA to JMJD6 (#63) | Dharmacon | CCAAAGUUAUCAAGGAAA | |
| Sequence-based reagent | siRNA to JMJD6 (#75) | Dharmacon | CAGUGAAGAUGAAGAUGAA | |
| Sequence-based reagent | siRNA to U2AF2 (#1) | Dharmacon | AGAAGAAGAAGGUCCGU | |
| Sequence-based reagent | siRNA to U2AF2 (#2) | Dharmacon | GUGGCAGUUUCAUAUUUG | |
| Sequence-based reagent | siRNA to CPSF6 (#1) | Dharmacon | GGAUCACCUUCCAAGACA | |
| Sequence-based reagent | siRNA to CPSF6 (#2) | Dharmacon | AGAACCGUCAUGACGAUU | |
| Sequence-based reagent | siRNA to GLS | Dharmacon | CAACTGGCCAAATTCAGTC | |
| Sequence-based reagent | siRNA to GLS (GAC-specific isoform) | Dharmacon | CCTCTGTTCTGTCAGAGTT | |
| Sequence-based reagent | siRNA to GLS (KGA-specific isoform) | Dharmacon | ACAGCGGGACTATGATTCT | |
| Sequence-based reagent | gDNA | Synthego | GGACTCTGGAGCGCCTAAAA | |
| Software, algorithm | CRIS.py | https://github.com/patrickc01/CRIS.py; Connelly and Pruett-Miller, 2019 | CRISPR-editing analysis software | |
| Software, algorithm | Sequest (version 28 revision 13) | Mark P Jedrychowski, et al. | Database search algorithm for mass spectrometry-based protein detection | |
| Software, algorithm | JUMPm | St Jude | Metabolomics data analysis software | |
| Software, algorithm | MoNA | https://mona.fiehnlab.ucdavis.edu/ | MS/MS library, used for metabolomics | |
| Software, algorithm | Human Metabolome Database (HMDB) | https://hmdb.ca/ | MS/MS library, used for metabolomics | |
| Software, algorithm | mzCloud | https://mzcloud.org | MS/MS library, used for metabolomics | |
| Software, algorithm | Trim-Galore v0.60 | https://github.com/FelixKrueger/TrimGalore; Krueger, 2023 | Software used to trim raw reads | |
| Software, algorithm | STAR v2.7 | St Jude | Pipeline used to map RNA reads to human genome, and differential gene expression | |
| Software, algorithm | R limma package v3.42.2 | https://bioconductor.org/packages/release/bioc/html/limma.html | Software used to normalize and transform read counts | |
| Software, algorithm | Molecular Signatures Database (MSigDB 6.2) (gsea2 v2.2.3) | https://www.gsea-msigdb.org/gsea/index.jsp | Software used to perform gene set enrichment analysis (GSEA) | |
| Software, algorithm | rMATS v4.1.0 | https://rnaseq-mats.sourceforge.io/rmats4.1.0/download.html | Software used for RNA alternative splicing analysis using mapped BAM files as input | |
| Software, algorithm | R2 | https://hgserver1.amc.nl/cgi-bin/r2/main.cgi?open_page=login | Portal used to investigate expression of JMJD6, GAC, and KGA in tumor tissues | |
| Software, algorithm | Kocak dataset GSE45547 (649 samples) | https://hgserver1.amc.nl/cgi-bin/r2/main.cgi?open_page=login | Dataset used to investigate expression of JMJD6, GAC, and KGA in tumor tissues | |
| Software, algorithm | Fischer dataset GSE120572 (394 samples) | https://hgserver1.amc.nl/cgi-bin/r2/main.cgi?open_page=login | Dataset used to investigate expression of JMJD6, GAC, and KGA in tumor tissues | |
| Software, algorithm | St Jude cloud | https://pecan.stjude.cloud/ | Portal used to investigate expression of JMJD6, GAC, and KGA in pediatric tumors | |
| Software, algorithm | cBioportal | http://cbioportal.org | Portal used to investigate copy number alterations of JMJD6 and Kaplan-Meier analyses | |
| Software, algorithm | DepMap | https://depmap.org/portal/ | Portal used to investigate metabolite abundance | |
| Software, algorithm | STRING program | https://string-db.org | Software used for network interaction analysis | |
| Software, algorithm | Cytoscape | https://cytoscape.org/ | Software used for presenting network interactions | |
| Software, algorithm | CRAN | https://CRAN.R-project.org/package=pheatmap | CNV heatmap generation | |
| Software, algorithm | GraphPad Prism v9 | https://www.graphpad.com/ | Software used for statistical analysis | |
| Software, algorithm | ImageJ | https://imagej.net/ij/ | Software used for colony formation/density |
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/90993/elife-90993-mdarchecklist1-v1.docx
-
Source data 1
JMJD6 interactome in BE2C cells and related pathways.
- https://cdn.elifesciences.org/articles/90993/elife-90993-data1-v1.zip