Adenine methylation is very scarce in the Drosophila genome and not erased by the ten-eleven translocation dioxygenase
Figures
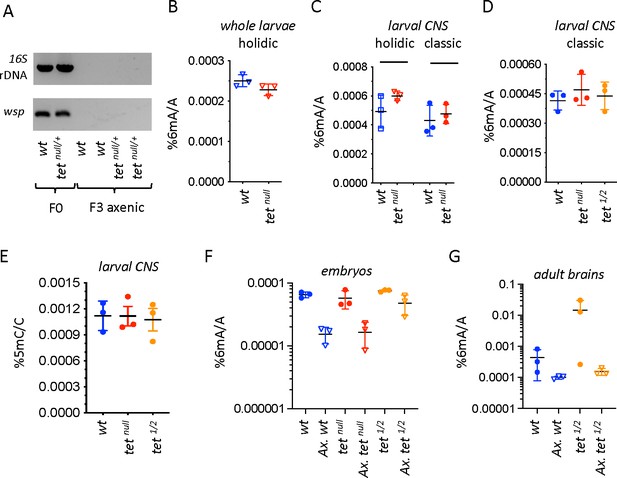
N6-methyladenine (6mA) levels in axenic Drosophila larvae are very low and not affected by ten-eleven translocation (TET) loss.
(A) The presence of bacterial contaminations in wild-type (wt) and tetnull/+ adult flies was checked by PCR using universal primers against bacterial 16 S rDNA and against the endosymbiotic bacteria gene wsp. PCRs were performed on DNA from parental (F0) flies and after three generations of breeding in axenic conditions (F3). (B–D) 6mA levels were measured by liquid chromatography with tandem mass spectrometry (LC-MS/MS) in genomic DNA (gDNA) from whole larvae (B) or dissected central nervous system (CNS) (C, D) generated from axenic flies reared on holidic medium (B, C) or conventional flies reared on classic medium (C, D). (E) 5-methylcytosine (5mC) levels were measured by LC-MS/MS in gDNA from dissected CNS from flies reared on classic medium. (F, G) 6mA levels were measured by LC-MS/MS in embryos (F) and dissected adult brains (G) collected from crosses with conventional or axenic (Ax.) individuals raised on classic fly medium supplemented (Ax.) or not with antibiotics. wt: wild-type (w1118); tetnull: tetnull/null; tet1/2: tetDMAD1/DMAD2. Filled circles: conventional flies; open triangles: axenic flies. Individual values, means and standard deviations are plotted. No statistically significant differences were observed between wt and tet mutant samples (Mann-Whitney test).
-
Figure 1—source data 1
Original file of the raw gels of PCR analyses for bacterial contaminations.
- https://cdn.elifesciences.org/articles/91655/elife-91655-fig1-data1-v1.zip
-
Figure 1—source data 2
PDF containing Figure 1A and raw gel pictures with relevant labels.
- https://cdn.elifesciences.org/articles/91655/elife-91655-fig1-data2-v1.zip
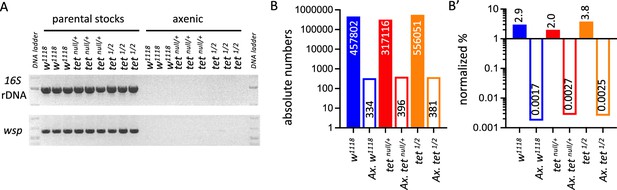
Detection of bacterial contamination in parental stocks and after three generations of breeding in axenic conditions.
(A) The presence of bacterial contamination in adult flies of the indicated genotypes was checked by PCR using universal primers against bacterial 16 S rDNA and against the endosymbiotic bacteria gene wsp. (B, B’) Genome-wide sequencing was used to assess the presence of contamination in genomic DNA (gDNA) from adult flies of the indicated genotypes. Between 15 and 19 million reads were analyzed per sample. The absolute numbers of reads mapping to bacteria, virus, or fungi genomes are represented in B, and their proportions normalized to the total number of reads are presented in B’. tet1/2: tetDMAD1/DMAD2.
-
Figure 1—figure supplement 1—source data 1
Original file of the raw gels of PCR analyses for bacterial contaminations.
- https://cdn.elifesciences.org/articles/91655/elife-91655-fig1-figsupp1-data1-v1.zip
-
Figure 1—figure supplement 1—source data 2
PDF containing Figure 1—figure supplement 1a and raw gel pictures with relevant labels.
- https://cdn.elifesciences.org/articles/91655/elife-91655-fig1-figsupp1-data2-v1.zip
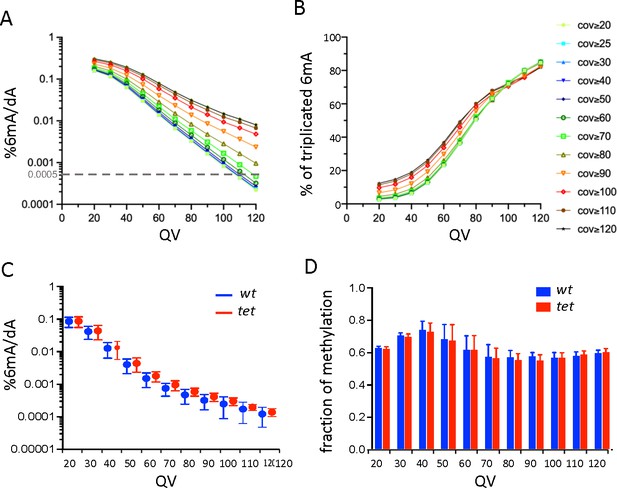
Single-molecule real-time sequencing (SMRT-seq) analysis of larval central nervous system (CNS) genomic DNA (gDNA) does not reveal an increase in N6-methyladenine (6mA) in the absence of ten-eleven translocation (TET).
(A) Percentage of adenines identified as 6mA in the wild-type fusion dataset depending on the quality value (QV) and coverage values used for 6mA selection. The dashed gray line indicates the level of 6mA measured by liquid chromatography with tandem mass spectrometry (LC-MS/MS) (0.0005%). (B) Influence of the QV and the coverage values on the proportion of 6mA identified in the wild-type fusion dataset and in the three original samples. (C) Percentage of adenines covered at least 25 x and identified as 6mA in each of the three wild-type (wt) or tet null (tet) datasets depending on the QV. Means and standard deviations are represented. (D) Fraction of methylation in wt or tet null datasets depending on the QV (coverage ≥25 x). Means and standard deviations are represented.
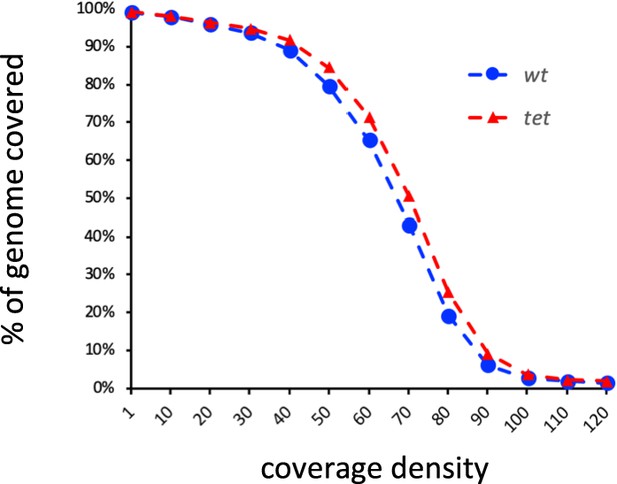
Proportion of the Drosophila genome covered in wild-type (wt) and tetnull (tet) single-molecule real-time sequencing (SMRT-seq) fusion datasets according to coverage density.
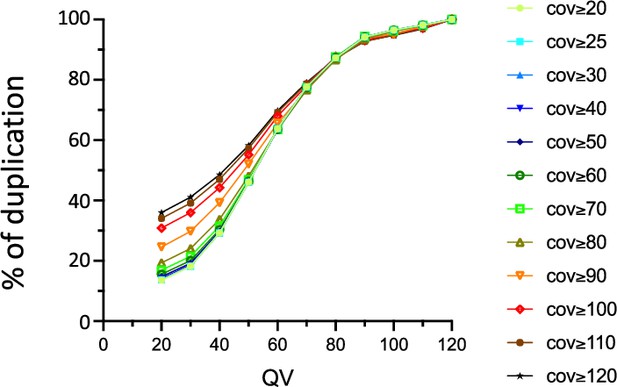
Influence of the quality value (QV) and the coverage values on the proportion of N6-methyladenine (6mA) identified in the wild-type (wt) fusion dataset and in the three original samples.
The percentage of 6mA identified in at least 2 out of 3 replicates is shown.
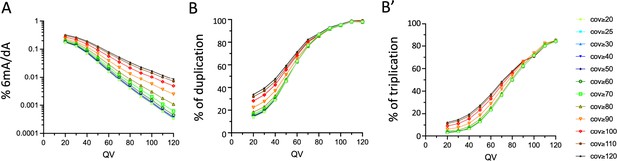
Influence of the quality and coverage values on SMRT-seq based detection of 6mA in the CNS of tetnull larvae.
(A) Percentage of all adenines identified as N6-methyladenine (6mA) by SMRT-seq in the tetnull fusion dataset depending on the quality value (QV) and coverage values used for 6mA selection (log10 scale). (B, B’) Influence of the QV and the coverage values on the proportion of 6mA identified in the tetnull fusion dataset and in the three original samples. (B) percentage of 6mA identified in at least 2 out of 3 samples; (B’): percentage of 6mA identified in all three samples.
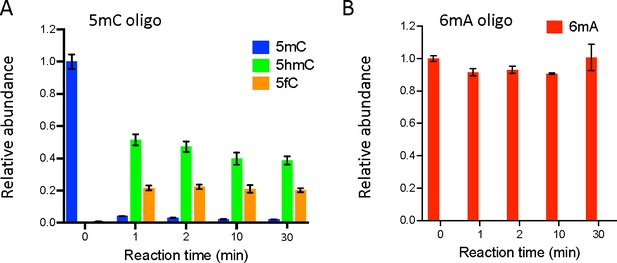
Ten-eleven translocation (TET) does not oxidize N6-methyladenine (6mA).
(A, B) In vitro assays showing TET activity profile on 5-methylcytosine (5mC) (A) and 6mA (B) containing double-stranded oligonucleotide substrates. The levels of 5mC and its oxidized products (5-hydroxymethylcytosine: 5hmC and 5fC) are represented relative to the 5mC level at t=0. The levels of 6mA are represented relative to 6mA level at t=0. Error bars denote standard deviations from three independent experiments.
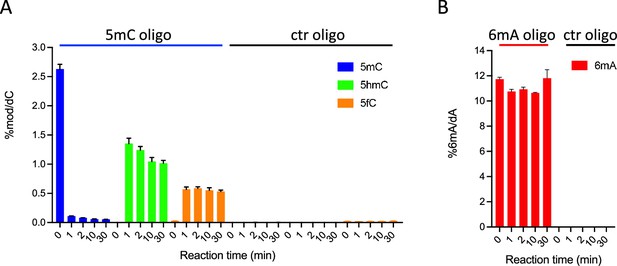
In vitro assays showing ten-eleven translocation (TET) activity profile on double-stranded oligonucleotide substrates containing or not 5-methylcytosine (5mC) (A) or N6-methyladenine (6mA) (B).
The levels of 5mC, 5-hydroxymethylcytosine (5hmC), and 5fC normalized to dC (A) or 6mA normalized to dA (B) are represented. Error bars denote standard deviations from three independent experiments. Only background levels of modified nucleosides were detected when purified recombinant TET catalytic domain was incubated with unmodified (ctr) oligonucleotides.

Multiple sequence alignment of ten-eleven translocation (TET)/JBP family members.
The sequence surrounding the HxD motif in Coprinopsis cinerea TET (ccTET), Trypanosoma brucei JBP1, Drosophila melanogaster TET (dTET), and Homo sapiens TET1, TET2, TET3 is shown. Conserved amino acids between TET homologs are boxed in yellow. The two key amino acids required for N6-methyladenine (6mA) oxidation by ccTET are labeled with a star.
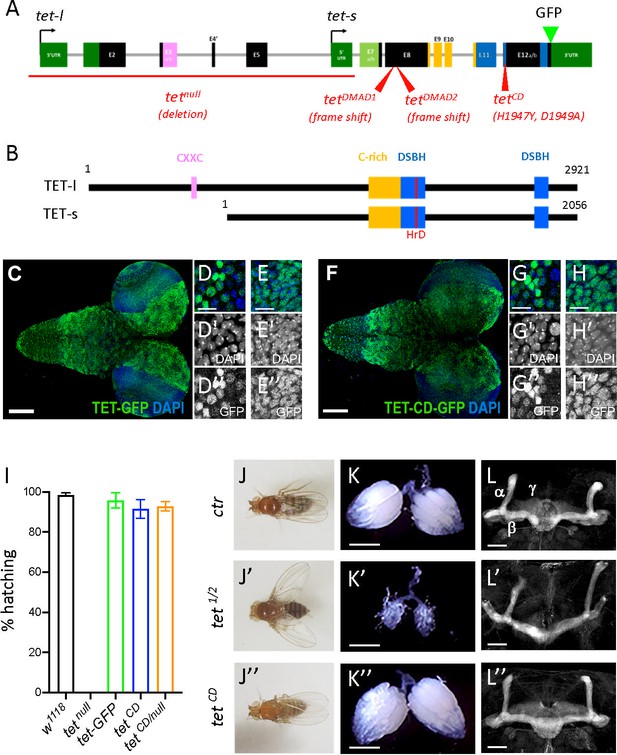
Ten-eleven translocation (TET) catalytic activity is largely dispensable in Drosophila.
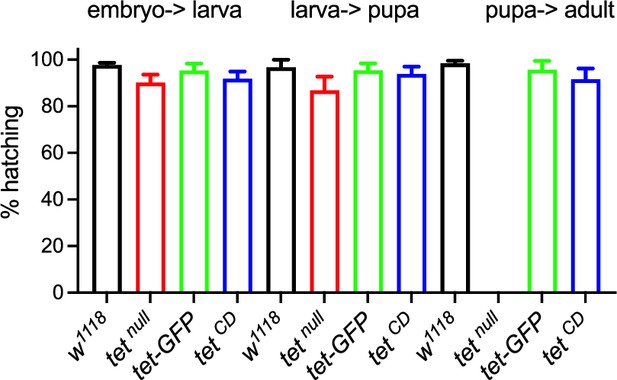
(A, B) Schematic representation of tet locus (A) and main protein isoforms (B). (A) tet is transcribed from two alternative promoters giving rise to tet-long (tet-l) and tet-short (tet-s) isoforms. Filled boxes represent exons; non-coding exons (UTR) are depicted in green, and coding exons in black or according to their domain-associated color. Introns are represented as gray lines (not to scale). The tet null, DMAD1, DMAD2, and catalytic dead (CD) alleles are depicted in red. The location of the GFP insertion generated by CRISRP/Cas9-mediated knock-in is also indicated. (B) The conserved domains of TET are colored; pink: CXXC DNA binding domain, orange: Cystein-rich domain, blue: double-stranded ß helix (DSBH) domain, red: HxD (iron binding motif). Amino acid positions are indicated according to the longest TET-l and TET-s isoforms. (C–H) Expression pattern of the wild-type and catalytic-dead versions of TET in the larval central nervous system (CNS). tet-GFP (C–E) and tetCD-GFP (F–H) knock-in lines were used to detect TET proteins by confocal imaging after immunostaining against GFP (green). Nuclei were labeled with DAPI (blue). (C, F): stitched images showing dorsal views of the entire CNS. Scale bar: 100 µm. (D, E, G, H): high-magnification views of TET expression in the ventral nerve cord (D, G) or the central brain (E, H). DAPI-only and GFP-only channels are presented in the middle (‘) and lower (‘’) panels, respectively. Scale bar: 10 µm. (i) Percentage of adult flies of the indicated genotypes hatching from their pupal case. Means and standard deviations from four independent experiments. (J–L) Wing positioning (J), ovaries (K), and mushroom bodies (L) of wild-type adult flies (ctr: tet-GFP) as compared to flies lacking TET expression (tet1/2: tetDMAD1/DMAD2 adult escapers) or TET enzymatic activity (tetCD). (L-L") Immunostaining against Fas2 on adult brains was used to label mushroom body α, β, and γ lobes. (K-K") Scale bar 500 µm. (L-L") Scale bar 50 µm.
Additional files
-
Supplementary file 1
Genome coverages of wild-type and tetnull fusin datasets in SMRT-seq.
- https://cdn.elifesciences.org/articles/91655/elife-91655-supp1-v1.xlsx
-
Supplementary file 2
Percentages of 6mA/A according to QV and coverage cut-off in wild-type and tetnull fusion datasets.
- https://cdn.elifesciences.org/articles/91655/elife-91655-supp2-v1.xlsx
-
Supplementary file 3
Proportions of replicated 6mA (triplicated and/or duplicated).
- https://cdn.elifesciences.org/articles/91655/elife-91655-supp3-v1.xlsx
-
Supplementary file 4
Percentages of 6mA/A and 6mA fractions of methylation according to QV in the three wild-type or three tetnull datasets (coverage ≥25 x).
- https://cdn.elifesciences.org/articles/91655/elife-91655-supp4-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/91655/elife-91655-mdarchecklist1-v1.docx