Expression of modified FcγRI enables myeloid cells to elicit robust tumor-specific cytotoxicity
Figures
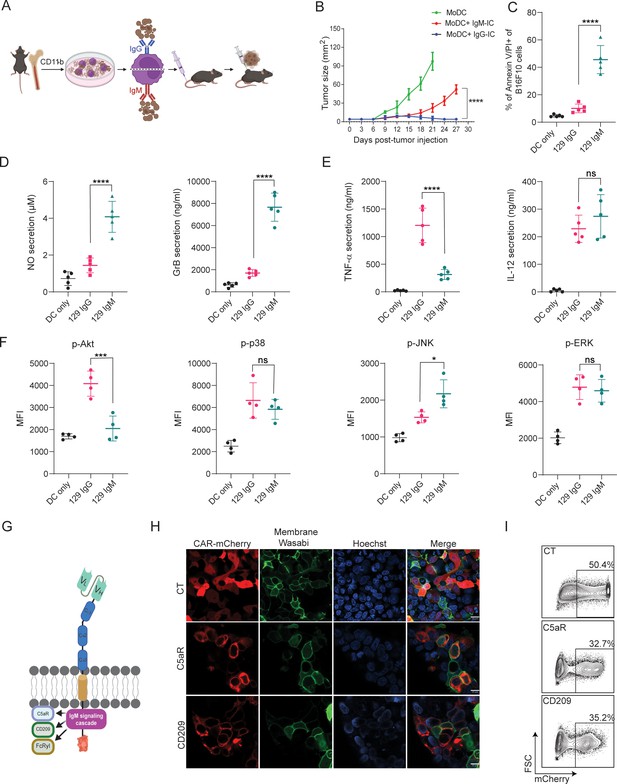
IgM-induced signaling elicits cytotoxic response in macrophages and can be integrated to a CAR design.
(A) Illustration of experimental setting. (B) B16F10 tumor size (mm2) in mice following prophylactic immunization with MoDC pulsed with tumor cells coated with allogeneic IgG or IgM (n=4). (C) Mean percentages of B16F10 melanoma cells stained for Annexin V/PI incubated with allogenic IgG and IgM following incubation with MoDC (n=5). (D–E) Mean levels of Granzyme B and NO (D) and proinflammatory cytokines (E) in the supernatants of MoDC following overnight activation with IgG and IgM immune complexes (n=5). (F) Mean fluorescent intensity (MFI) of MAPK enzymes in MoDC following activation for 20 min with IgG and IgM tumor immune complexes (n=5). (G) Illustration representing CAR-macrophage design. (H) Confocal microscopy images of HEK293FT cells 24 hr post-transfection with CAR plasmids and membranous wasabi. (I) Representative FACS analysis of HEK239FT cells 24 hr post-transfection with CAR plasmids. Results are from one representative experiment out of at least three performed. Statistical significance was calculated using non-parametric t-test (* denote p<0.05, *** denote p<0.001, **** denote p<0.0001).
© 2024, BioRender Inc. Figure 1A, G was created using BioRender, and is published under a CC BY-NC-ND license. Further reproductions must adhere to the terms of this license.
-
Figure 1—source data 1
IgM-induced signaling elicits cytotoxic response in macrophages and can be integrated to a CAR design.
(A) B16F10 tumor size (mm2) in mice following prophylactic immunization with MoDC pulsed with tumor cells coated with allogeneic IgG or IgM (n=4). (B) Percentage of B16F10 melanoma cells stained for Annexin V/PI incubated with allogenic IgG and IgM following incubation with MoDC (n=5).
- https://cdn.elifesciences.org/articles/91999/elife-91999-fig1-data1-v2.xlsx
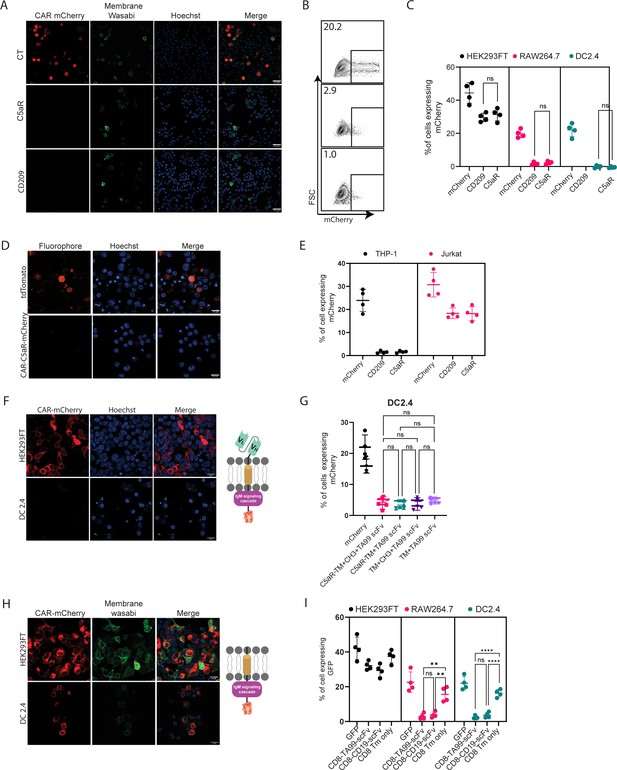
scFv is not expressed by myeloid cells.
(A) Confocal microscopy images of DC 2.4 cells 24 hr post-transfection with CAR plasmids and membranous wasabi. (B) Representative FACS analysis of DC 2.4 cells 24 hr post-transfection with CAR plasmids. (C) Percentages of transfected cells 24 hr post-transfection (n=4). (D) Confocal microscopy images of THP-1 cells 72 hr post lentiviral infection with CAR-C5aR-mCherry and tdTomato plasmids. (E) Percentages of transfected human cell lines 72 hr following transduction (n=4). (F–G) Representative confocal microscopy (F) and mean percentages (G) of cells expressing chimeric molecules 24 hr after transfection. (H–I) Representative confocal microscopy (H) and mean percentages of cells (I) expressing chimeric molecules 24 hr following transfection (n=4). Results are from one representative experiment out of at least three performed. Statistical significance was calculated using non-parametric t-test (** denote p<0.01, **** denote p<0.0001).
© 2024, BioRender Inc. Figure 2F, H was created using BioRender, and is published under a CC BY-NC-ND license. Further reproductions must adhere to the terms of this license.
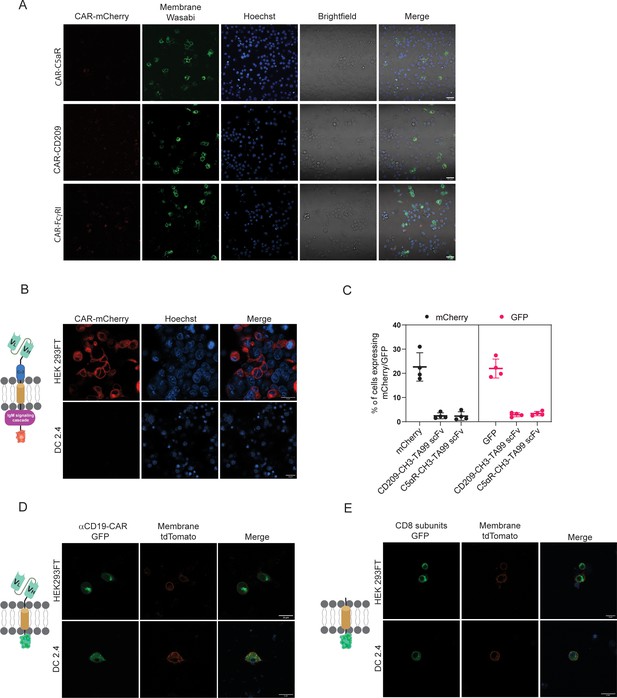
scFv is not expressed by myeloid cells.
(A) Confocal microscopy images of RAW264.7 cells transfected with scFv-based chimeric receptors. (B–C) Confocal microscopy images (B) and mean expression percentages (C) of HEK293FT and DC2.4 cells 24 hr post-transfection with TA99 scFv (n=4). (D–E) Confocal microscopy images of HEK293FT and DC 2.4 cells 24 hr post-transfection with αCD19 scFv (D) and with CD8-transmembrane portion only (E). Results are from one representative experiment out of at least three performed. Statistical significance was calculated using non-parametric t test.
© 2024, BioRender Inc. Figure 2—figure supplement 1B, D and E was created using BioRender, and is published under a CC BY-NC-ND license. Further reproductions must adhere to the terms of this license.
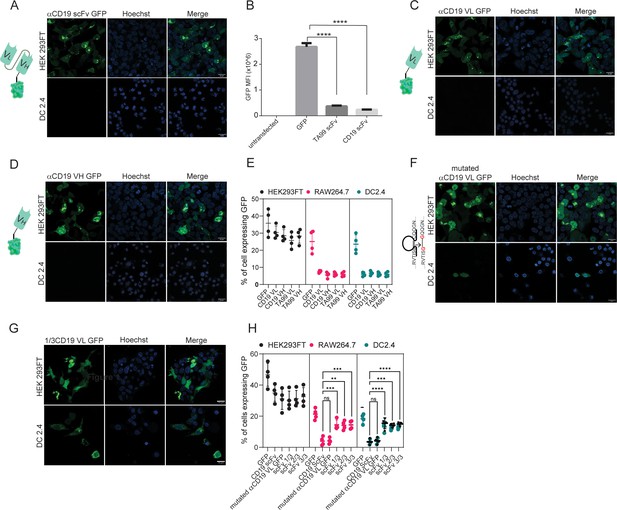
Both VH and VL domains prevent expression of ScFv in myeloid cells.
(A) Confocal microscopy images of DC 2.4 cells 24 hr post-transfection with αCD19-scFv GFP plasmid. (B) Geometric mean of GFP-positive cells 24 hr post-transfection with different αCD19- and TA99- ScFv GFP constructs in DC2.4 (n=3). (C) Confocal microscopy images of HEK 293 FT and DC 2.4 cells 24 hr post-transfection with αCD19-variable light chain GFP plasmid. (D) Confocal microscopy images of HEK 293 FT and DC 2.4 cells 24 hr post-transfection with αCD19-variable heavy chain GFP plasmid. (E) Mean percentages of cells expressing scFv fragments 24 hr post-transfection (n=4). (F) Left: Illustration of mutated variable light chain. Right: Confocal microscopy images of HEK 293 FT and DC 2.4 cells 24 hr post-transfection with αCD19- mutated (linear) variable light chain. (G) Confocal microscopy images of HEK293FT and DC2.4 cells 24 hr post-transfection with 1/3 fragments of αCD19-variable light chain GFP plasmid. (H) Mean percentages of GFP-positive cells 24 hr post-transfection with different fragments of scFv-GFP in DC2.4 and RAW264.7 (n=4). Results are from one representative experiment out of at least three performed. Statistical significance was calculated using non-parametric t test (** denote p<0.01, *** denote p<0.001, **** denote p<0.0001).
© 2024, BioRender Inc. Figure 3A, C and D was created using BioRender, and is published under a CC BY-NC-ND license. Further reproductions must adhere to the terms of this license.
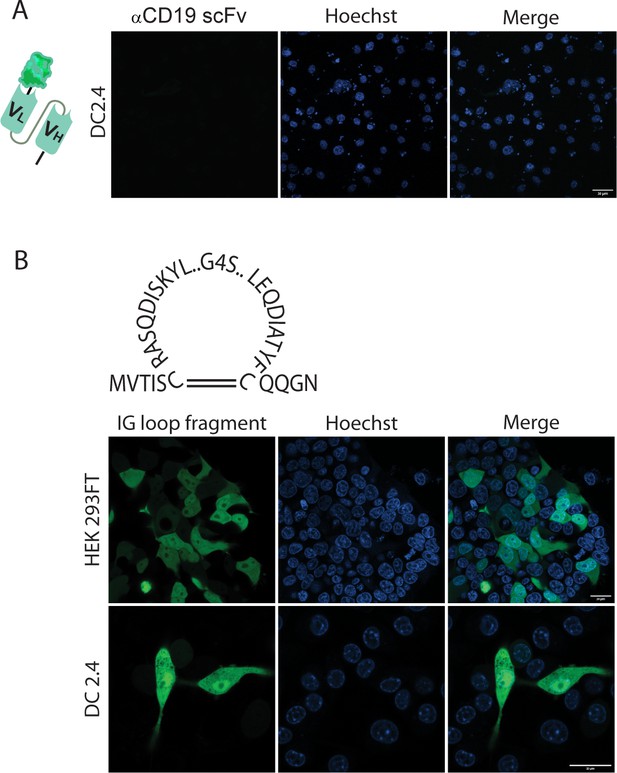
Immunoglobulin structure of scFv does not prevent degradation by myeloid cells.
(A) Representative confocal images of DC 2.4 transfected with αCD19 scFv fused to GFP at the carboxyl end. (B) Upper: Illustration of immunoglobulin loop fragment GFP. Lower: Confocal microscopy images of HEK293FT and DC2.4 24 hr post-transfection with immunoglobulin loop fragment. Results are from one representative experiment out of at least three performed.
© 2024, BioRender Inc. Figure 3—figure supplement 1A was created using BioRender, and is published under a CC BY-NC-ND license. Further reproductions must adhere to the terms of this license.
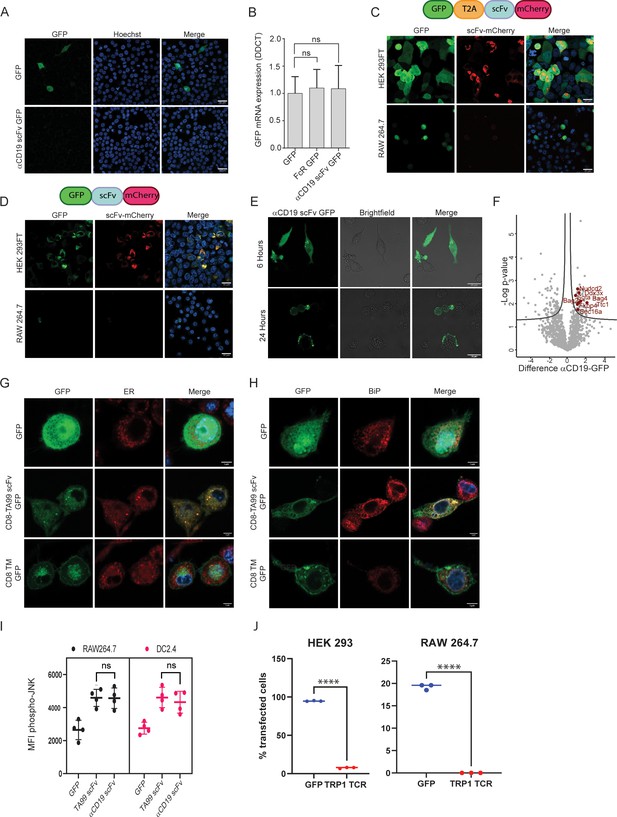
scFv fragments induce ER stress in myeloid cells.
(A) Confocal microscopy imaging of RAW 264.7 24 hr post-transfection with linear mRNA vectors translating to GFP and αCD19-scFv GFP. (B) qPCR data showing relative mRNA levels in RAW 264.7 transfected with GFP, Fc receptor-GFP, and αCD19-scFv GFP (n=4). (C) Upper: Illustration of plasmid subunits. Lower: Confocal microscopy imaging of HEK293FT and RAW 264.7 24 hr post-transfection with T2A ribosomal skipping plasmid including ScFv. (D) Upper: Illustration of plasmid subunits. Lower: Confocal microscopy imaging of HEK293FT and RAW 264.7 24 hr post-transfection with plasmid not containing T2A. (E) Confocal microscopy images of RAW264.7 cells at 6 hr and 24 hr post-transfection with αCD19-ScFv GFP plasmid. (F) Volcano plot showing differentially expressed proteins αCD19 ScFv GFP and GFP in DC 2.4 cells. (G) Confocal microscopy images of DC 2.4 stained with an ER stain, 24 hr post-transfection with GFP, membranous TA99-ScFv GFP. (H) Confocal microscopy images of DC 2.4 stained for BiP 24 hr post-transfection. (I) Mean levels of phospho-JNK 6 hours following transfection (n=4). (J) Percentage of cells expressing GFP or TRP-TCR1 24 hr following transfection (n=3). Results are from one representative experiment out of at least three performed. Statistical significance was calculated using non-parametric t-test (**** denote p<0.0001).
-
Figure 4—source data 1
scFv fragments induce ER stress in myeloid cells.
(A) qPCR data showing mRNA levels in RAW 264.7 transfected with GFP, Fc receptor-GFP, and αCD19-ScFv GFP (n=4). (B) LC-MS data results showing protein complexes pulled down using GFP magnetic beads in DC 2.4 6 hr post transfection with αCD19 ScFv GFP and GFP. (C) Flow cytometry data of CFSE dilution in CD8+ and CD4+ T cells following co-culture with ova conjugated scFv.
- https://cdn.elifesciences.org/articles/91999/elife-91999-fig4-data1-v2.xlsx
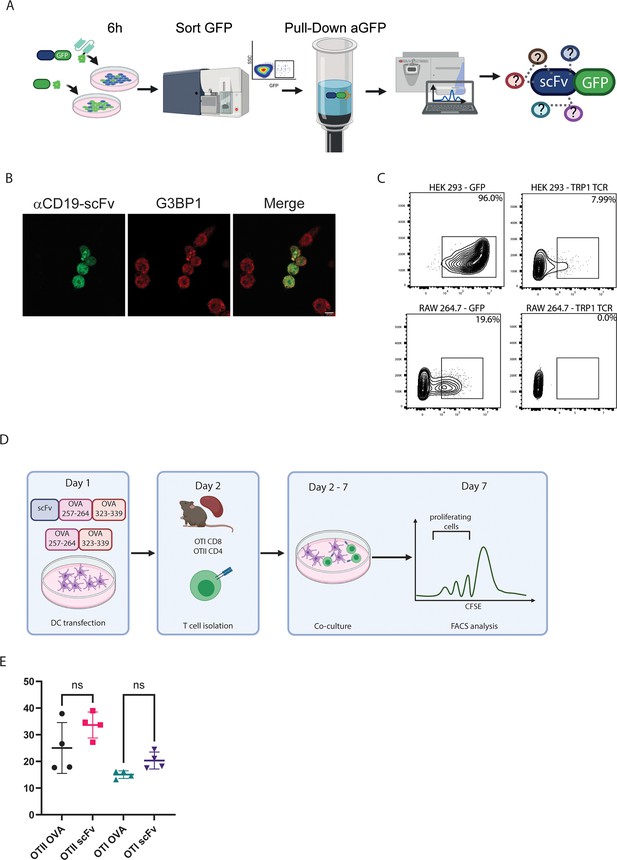
scFv induces ER stress response in myeloid cells.
(A) Iillustration of experimental design. (B) Confocal microscopy images with staining for G3BP1 in DC2.4 cells 6 hr post-transfection with αCD19 scFv. (C) Representative FACS analysis of HEK239FT and RAW 264.7 cells 24 hr post-transfection with TRP1-TCR. (D) Illustration of experimental design. (E) Representative analysis of CFSE dilution in CD8+ and CD4+ T cells following co-culture with ova conjugated scFv (n=4). Results are from one representative experiment out of at least three performed. Statistical significance was calculated using non-parametric t-test.
© 2024, BioRender Inc. Figure 4—figure supplement 1A, D was created using BioRender, and is published under a CC BY-NC-ND license. Further reproductions must adhere to the terms of this license.
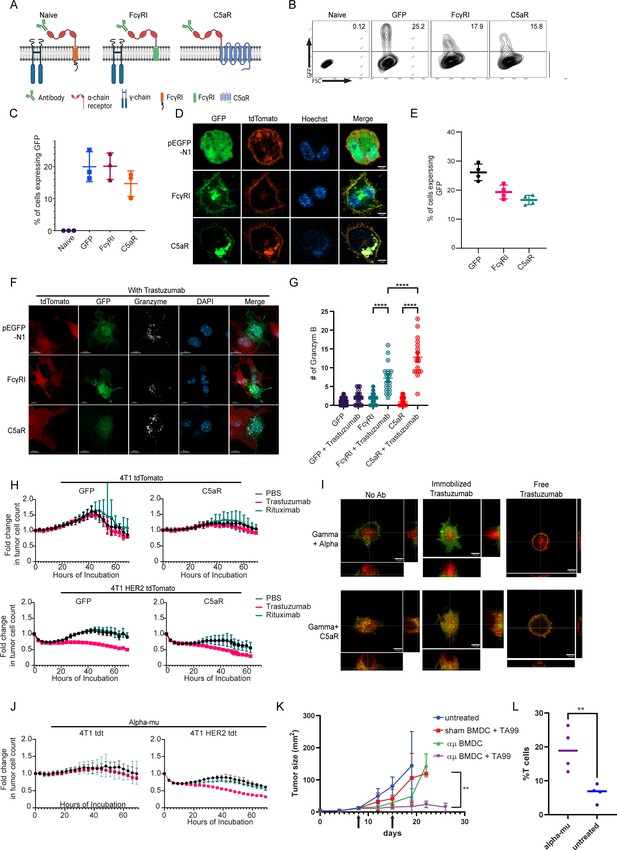
FcγRI can provide a scaffold for incorporating IgM-induced signaling in myeloid cells and endows them with tumor cell-specific killing ability.
(A) Illustration of chimeric Fcγreceptor design. (B–C) Representative FACS plots (B) and mean percentages (C) of RAW 264.7 cells expressing chimeric Fcγ receptors 24 hr after transfection (n=3). (D) Confocal microscopy images of RAW 264.7 cells 24 hr post-transfection with Fcγ receptors tagged with GFP and membrane-tagged tdTomato. (E) Mean percentages of BMDC 72 hr post lentivirus transduction with Fcγ receptors (n=4). (F) Confocal microscopy staining of GrB in RAW 264.7 cells co-cultured overnight with 4T1 cells expressing human HER2. (G) Mean counts of GrB in the synapse between transduced RAW 264.7 cells and the tumor cells (n=18). (H) IncuCyte analysis of human HER2+ 4T1 cells growth following incubation with transduced RAW 264.7 cells (n=6). (I) Super-resolution microscopy of GFP-tagged chimeric FcγR and mCherry-tagged gamma chain. (J) IncuCyte analysis of human HER2+ 4T1 cells growth following incubation with transduced RAW 264.7 cells (n=6). (K) Tumor size measurements (mm2) in mice treated with mRNA transfected BMDC with and without antibody. Arrows point to subcutaneous injection of treated BMDC (n=5). (L) Mean percentages of CD3+ out of CD45+ cells in B16F10 tumors from day 26 (n=4). Results are from one representative experiment out of at least three performed. Statistical significance was calculated using non-parametric t test (** denote p<0.01, **** denote p<0.0001).
© 2024, BioRender Inc. Figure 5A was created using BioRender, and is published under a CC BY-NC-ND license. Further reproductions must adhere to the terms of this license.
-
Figure 5—source data 1
FcγRI can provide a scaffold for incorporating IgM-induced signaling in myeloid cells and endows them with tumor cell-specific killing ability.
(A) Tumor size measurements (mm2) in mice treated with mRNA transfected BMDC with and without antibody (n=6). (B) Mean percentages of CD3+ out of CD45+ cells in B16F10 tumors from day 26 (n=4).
- https://cdn.elifesciences.org/articles/91999/elife-91999-fig5-data1-v2.xlsx
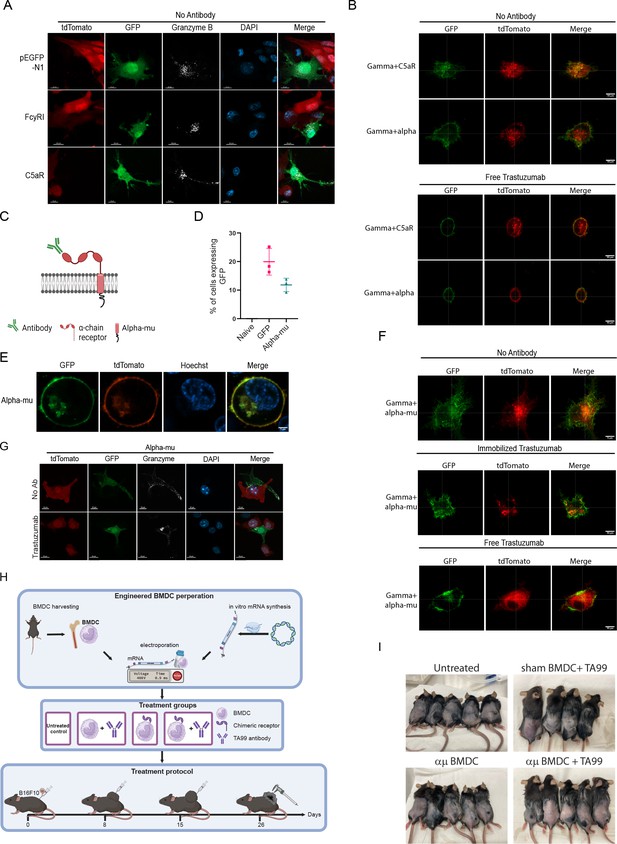
FcγRI can be used as a scaffold to transmit IgM-induced signaling.
(A) Representative confocal staining of Granzyme B in transduced RAW 264.7 cells incubated overnight with 4T1 cells expressing human HER2 antigen. (B) Super-resolution microscopy of RAW 264.7 cells transfected with chimeric FcγRI molecules as such, or one hour after addition of free antibody. (C) Illustration of receptor design. (D) Mean percentages of RAW 264.7 cells expressing chimeric receptor (n=4). (E) Representative microscopy of RAW 264.7 cells co-transfected with GFP-fused chimeric receptor and mCherry-membrane protein. (F) Super-resolution microscopy of RAW 264.7 cells transfected with chimeric FcγRI molecules as such, or one hour after addition of free antibodies. (G) Confocal microscopy staining of GrB in RAW 264.7 cells co-cultured overnight with 4T1 cells expressing human HER2. (H) Illustration of experimental setting. (I) Representative photomicrographs of tumor-bearing mice 26 days after tumor injection. Results are from one representative experiment out of at least three independent experiments performed.
© 2024, BioRender Inc. Figure 5—figure supplement 1C, H was created using BioRender, and is published under a CC BY-NC-ND license. Further reproductions must adhere to the terms of this license.
Tables
scFv construct design and preparation process.
| Construct subunits design | Construct preparation |
|---|---|
| (ssCD8α) – TA99 scFv – CH1 – hinge – CH2 – CH3 – transmembrane +intracellular portion of CD209 /C5aR−mCherry | All subunits were designed and designed and ordered in gBlock format from Integrated DNA Technologies (IDT) (Coralville, IA, USA). Subunits were added to pcDNA 3.1 (+) using Gibson assembly, and cloned to pmCherry- N1. |
| (ssCD8α) – TA99 scFv –– CH3 – transmembrane +intracellular portion of CD209 /C5aR- mCherry | Construct was created by elimination of CH1-hinge-CH2 subunits through invert PCR followed by kinase, ligase, DpnI (KLD) enzyme mix. |
| pLVX: TA99 scFv – CH3 – (TM +IC) C5aR - mCherry | pLVX-IRES-Hyg vector was restricted using SpeI/NotI. Receptor insert including TA99 scFv - heavy chain - C5aR (transmembrane +intracellular) - mCherry was restricted from mCherry vector via restriction enzymes NheI/NotI followed by ligation using 4T DNA ligase. |
| (ssCD8α) – CH1 – hinge – CH2 – CH3 – transmembrane +intracellular portion of CD209 /C5aR- mCherry | Constant heavy sequence was isolated and amplified using invert PCR for scFv removal followed by KLD enzymes. |
| (ssCD8α) – TA99 scFv - transmembrane and intracellular portion of either CD209 or C5aR- mCherry | Removal of constant heavy subunits was done by invert PCR amplification. Followed by KLD enzyme reaction. |
| (ssCD8α) – TA99 scFv – CD8α hinge – CD8α transmembrane – mCherry | Extracellular portion was isolated via restriction with BamHI/EcoRI and inserted into pmCherryN1 vector followed by ligation with 4T DNA ligase. |
| (ssCD8α) –CD8α hinge - a transmembrane and intracellular portion of either CD209 or C5aR - mCherry | scFv portion was removed via invert PCR using Fw primer that included a tail of CD8α hinge sequence as an extracellular remnant. The PCR product was used in KLD enzymes mix reaction. |
| TA99 scFv - GFP | TA99 scFv sequence was restricted from pemCherry-N1 plasmid using EcoRI/XhoI enzymes and inserted to peGFP-N1 plasmid linearized with same enzymes. |
| GFP - T2A - TA99 scFv - mCherry | T2A sequence was designed and ordered in linear gBlock sequence, flanked by restriction enzymes: BglII/NheI. The sequence was inserted to peGFP-C1 vector. GFP-T2A sequence was isolated via restriction with NheI/XhoI and inserted to plasmid which included an insert of TA99 scFv – mCherry. |
| GFP – TA99 scFv – mCherry | Removal of T2A sequence was performed by invert PCR followed by KLD enzyme mix reaction. |
| (ssCD8α) – αCD19 scFv – CD8α hinge – CD8α transmembrane – mCherry | (ssCD8α) – TA99 scFv – CD8α hinge – CD8α transmembrane – mCherry was used as backbone. TA99was removed by invert PCR amplification exclusion ofαCD19 scFv was amplified using plasmid pHR-PGK-antiCD19-synNotch-GalVP64 (plasmid#79125) which was purchased from Addgene (Watertown, MA, USA). Both backbone and insert were flanked with XhoI/EcoRI. |
| αCD19 scFv - GFP | αCD19 scFv from plasmid#79125 (Addgene) was amplified using primers with tails encoding restriction sites for XhoI/EcoRI enzymes. peGFP-N1 backbone was restricted using same enzymes. |
| pGEM4Z: T7 promoter - αCD19 scFv – GFP – polyA tail | αCD19 scFv was cloned into pGEM4Z using restriction enzymes NheI +NotI. pGEM4Z GFP was restricted using: XbaI +NheI. |
| pLVX: αCD19 scFv - GFP | αCD19 – GFP was isolated from peGFP-N1 backbone using enzymes NheI/NotI and inserted into pLVX backbone linearized using SpeI/NotI. |
| GFP - αCD19 scFv | αCD19 scFv sequence was isolated and amplified using primers which included overhang sequence of restriction sites for enzymes XhoI/EcoRI. peGFPC1 was restricted with same enzymes followed by ligation. |
| TA99 variable heavy chain-GFP | Variable heavy chain was isolated and amplified using invert PCR for variable light chin removal followed by KLD enzyme mix. |
| TA99 variable light chain-GFP | Variable light chain was isolated and amplified using invert PCR for variable heavy chin removal followed by KLD enzyme mix. |
| αCD19 variable heavy chain-GFP | Variable heavy chain was isolated and amplified using invert PCR for variable light chin removal followed by KLD enzyme mix. |
| αCD19 variable light chain-GFP | Variable light chain was isolated and amplified using invert PCR for variable heavy chin removal followed by KLD enzyme mix. |
| αCD19 mutated VL (linear) | G-block of mutated αCD19 variable light chain. Two-point mutations included replacement of Cysteine amino acid with glycine in positions 24 and 89, flanked by restriction enzymes XhoI/EcoRI was designed and orders from IDT. PeGFP-N1 plasmid was restricted with same enzymes. |
| αCD19 mutated VH (linear) | gBlock of mutated αCD19 variable heavy chain. Two-point mutations included replacement of Cysteine amino acid with glycine in positions 22 and 95, flanked by restriction enzymes XhoI/EcoRI was designed and orders from IDT. PeGFP-N1 plasmid was restricted with same enzymes. |
| Immunoglobulin loop fragment GFP | DNA sequence was ordered in linear gBlock from IDT. Insert of interest was flanked by XhoI/EcoRI and inserted into peGFP-N1 backbone linearized with both enzymes. Final amino acid sequence included: MVTISCRASQDISKYL-GGGGSGGGGSGGGGS-EQEDIATYFCQQGN |
| (ssCD8α) – CD64 – transmembrane +intracellular portion of C5aR/CD209/μ – GFP | CD64 sequence was purchased in gBlock format from IDT, subunits were added in sequence keeping reading frame in check using homology sequences into peGFP-N1. |
| (ssCD8α) – CD64 – transmembrane +intracellular portion of C5aR/CD209/μ – T2A - GFP | T2A sequence was ordered in linearized DNA gBlock from IDT. Sequence was flanked by homology sequence which overlapped linearized plasmid ends created by restriction with enzymes BamHI/XmaI. |
| pLVX: (ssCD8α) – CD64 – transmembrane +intracellular portion of C5aR /CD209 – T2A GFP | Receptor sequence including all subunits - GFP was isolated from peGFPN1 plasmid using restriction enzymes NheI/NotI. pLVX backbone was linearized using SpeI/NotI. |
| CD64 (extracellular +transmembrane) – GFP (used for RT PCR expression control) | Transmembrane +intracellular sequences were removed from (ssCD8α) – CD64 – transmembrane +intracellular sequence – GFP using invert PCR. Followed by KLD enzyme’s reaction. |
| Alpha mu in βGlobin5'UTR | Plasmid was ordered with insert in place through A2S technologies Ltd (Yavne, Israel) |
| anti TRP1 murine TCR | Minigene was cloned to pcDNA3-EGFP backbone plasmid using restriction enzymes. |
| TA99 scFv – OVA257-264 – G4S linker - OVA323-339 | DNA sequence including all subunits was designed and ordered in gBlock via IDT. Sequence was inserted into pcDNA 3.1 (+) using restriction enzymes. |
| OVA257-264 – G4S linker - OVA323-339 | Control construct to scFv-OVA was prepared by removal of scFv sequence using invert PCR amplification followed by a KLD reaction according to protocol. |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (57BL/6) | Wild-type (WT) C57BL/6 mice | Envigo | Strain #:000664 | |
| Strain, strain background (129S1) | 129S1/SvImJ | The Jackson Laboratory | Strain #:002448 | |
| Strain, strain background (OT-I) | C57BL/6-Tg(TcraTcrb)1100Mjb/J OT-I | The Jackson Laboratory | Strain #:003831 | |
| Strain, strain background (CD45.1) | B6.SJL-Ptprca Pepcb/BoyJ | The Jackson Laboratory | Strain #:002014 | |
| Strain, strain background (OT-II) | B6.Cg-Tg(TcraTcrb)425Cbn/J OT-II | The Jackson Laboratory | Strain #:004194 | |
| Cell line (Homo-sapiens) | Human Embryonic Kidney (HEK)–293 FT cells | ThermoFisher Scientific | R70007 | |
| Cell line (Mus musculus) | DC 2.4 | Merck Milipore | SCC142 | |
| Cell line (Mus musculus) | RAW 264.7 | ATCC | TIB-71 | |
| Cell line (Homo-sapiens) | Jurkat | ATCC | TIB-152 | |
| Cell line (Homo-sapiens) | THP-1 | ATCC | TIB-202 | |
| Cell line (Mus musculus) | B16F10 | ATCC | CRL-6475 | |
| Cell line (Mus musculus) | 4T1 | ATCC | CRL-2539 | |
| Transfected construct (4T1, Mus musculus) | 4T1 coexpressing H2b-Tdt / HER2 | This paper | Cell line was created using lentiviral vectors | |
| Antibody | anti-mouse CD64 (FcγRI) (mouse monoclonal) | Biolegend | Cat# 139315 clone: X54-5/7.1 | 1 µl in 100 µl volume |
| Antibody | anti-mouse CD3 (rat monoclonal) | Biolegend | Cat# 100201 Clone: 17-A2 | 0.5 µl in 100 µl volume |
| Antibody | anti-mouse CD45 (rat monoclonal) | Biolegend | Cat# 103101 Clone: 30-F11 | 0.5 µl in 100 µl volume |
| Antibody | Fluorochrome-conjugated antibodies against phospho-p38 (Thr180/Tyr182) (rabbit monoclonal) | Cell Signaling Technologies | Cat# 8623 | Dilution 1:50 |
| Antibody | Fluorochrome-conjugated antibodies against phospho-JNK (Ser63) (rabbit monoclonal) | Cell Signaling Technologies | Cat# 91952 | 1:100 |
| Antibody | phospho-ERK1/2 (p44) (pT202/pY204) (rabbit monoclonal) | Cell Signaling Technologies | Cat# 14095 | 1:50 |
| Antibody | phospho-Akt (pY473) (rabbit polyclonal) | Cell Signaling Technologies | Cat# 9271 | 1:100 |
| Recombinant DNA reagent | (ssCD8α) – TA99 scFv – CH1 – hinge – CH2 – CH3 – transmembrane +intracellular portion of CD209 /C5aR-mCherry | This paper | This construct was prepared in lab using linear DNA ordered from Integrated DNA Technologies (IDT). Detailed construct preparation can be found in Table 1 in main article file. | |
| Recombinant DNA reagent | (ssCD8α) – TA99 scFv –– CH3 – transmembrane +intracellular portion of CD209 /C5aR- mCherry | This paper | This construct was prepared in lab. Detailed construct preparation can be found in Table 1 in main article file. | |
| Recombinant DNA reagent | pLVX: TA99 scFv – CH3 – (TM +IC) C5aR - mCherry | This paper | This construct was prepared in lab. Detailed construct preparation can be found in Table 1 in main article file. | |
| Recombinant DNA reagent | (ssCD8α) – CH1 – hinge – CH2 – CH3 – transmembrane +intracellular portion of CD209 /C5aR- mCherry | This paper | This construct was prepared in lab. Detailed construct preparation can be found in Table 1 in main article file. | |
| Recombinant DNA reagent | (ssCD8α) – TA99 scFv - transmembrane and intracellular portion of either CD209 or C5aR- mCherry | This paper | This construct was prepared in lab. Detailed construct preparation can be found in Table 1 in main article file. | |
| Recombinant DNA reagent | (ssCD8α) – TA99 scFv – CD8α hinge – CD8α transmembrane – mCherry | This paper | This construct was prepared in lab. Detailed construct preparation can be found in Table 1 in main article file. | |
| Recombinant DNA reagent | (ssCD8α) –CD8α hinge - a transmembrane and intracellular portion of either CD209 or C5aR - mCherry | This paper | This construct was prepared in lab. Detailed construct preparation can be found in Table 1 in main article file. | |
| Recombinant DNA reagent | TA99 scFv - GFP | This paper | This construct was prepared in lab. Detailed construct preparation can be found in Table 1 in main article file. | |
| Recombinant DNA reagent | GFP - T2A - TA99 scFv - mCherry | This paper | This construct was prepared in lab using linear DNA ordered from (IDT). Detailed construct preparation can be found in Table 1 in main article file. | |
| Recombinant DNA reagent | GFP – TA99 scFv – mCherry | This paper | This construct was prepared in lab. Detailed construct preparation can be found in Table 1 in main article file. | |
| Recombinant DNA reagent | (ssCD8α) – αCD19 scFv – CD8α hinge – CD8α transmembrane – mCherry | This paper | This construct was prepared in lab. Detailed construct preparation can be found in Table 1 in main article file. | |
| Recombinant DNA reagent | αCD19 scFv - GFP | This paper | This construct was prepared in lab. Detailed construct preparation can be found in Table 1 in main article file. | |
| Recombinant DNA reagent | pGEM4Z: T7 promoter - αCD19 scFv – GFP – polyA tail | This paper | This construct was prepared in lab. Detailed construct preparation can be found in Table 1 in main article file. | |
| Recombinant DNA reagent | pLVX: αCD19 scFv - GFP | This paper | This construct was prepared in lab. Detailed construct preparation can be found in Table 1 in main article file. | |
| Recombinant DNA reagent | GFP - αCD19 scFv | This paper | This construct was prepared in lab. Detailed construct preparation can be found in Table 1 in main article file. | |
| Recombinant DNA reagent | TA99 variable heavy chain-GFP | This paper | This construct was prepared in lab. Detailed construct preparation can be found in Table 1 in main article file. | |
| Recombinant DNA reagent | TA99 variable light chain-GFP | This paper | This construct was prepared in lab. Detailed construct preparation can be found in Table 1 in main article file. | |
| Recombinant DNA reagent | αCD19 variable heavy chain-GFP | This paper | This construct was prepared in lab. Detailed construct preparation can be found in Table 1 in main article file. | |
| Recombinant DNA reagent | αCD19 variable light chain-GFP | This paper | This construct was prepared in lab. Detailed construct preparation can be found in Table 1 in main article file. | |
| Recombinant DNA reagent | αCD19 mutated VL (linear) | This paper | This construct was prepared in lab using linear DNA ordered from (IDT). Detailed construct preparation can be found in Table 1 in main article file. | |
| Recombinant DNA reagent | αCD19 mutated VH (linear) | This paper | This construct was prepared in lab using linear DNA ordered from (IDT). Detailed construct preparation can be found in Table 1 in main article file. | |
| Recombinant DNA reagent | Immunoglobulin loop fragment GFP | This paper | This construct was prepared in lab using linear DNA ordered from (IDT). Detailed construct preparation can be found in Table 1 in main article file. | |
| Recombinant DNA reagent | (ssCD8α) – CD64 – transmembrane +intracellular portion of C5aR/CD209/μ – GFP | This paper | This construct was prepared in lab using linear DNA ordered from (IDT). Detailed construct preparation can be found in Table 1 in main article file. | |
| Recombinant DNA reagent | (ssCD8α) – CD64 – transmembrane +intracellular portion of C5aR/CD209/μ – T2A - GFP | This paper | This construct was prepared in lab using linear DNA ordered from (IDT). Detailed construct preparation can be found in Table 1 in main article file. | |
| Recombinant DNA reagent | pLVX: (ssCD8α) – CD64 – transmembrane +intracellular portion of C5aR /CD209 – T2A GFP | This paper | This construct was prepared in lab. Detailed construct preparation can be found in Table 1 in main article file. | |
| Recombinant DNA reagent | CD64 (extracellular +transmembrane) – GFP | This paper | This construct was prepared in lab. Detailed construct preparation can be found in Table 1 in main article file. | |
| Recombinant DNA reagent | Alpha mu in βGlobin5'UTR | This paper | This construct was designed in lab and ordered in plasmid from A2S technologies | |
| Recombinant DNA reagent | anti TRP1 murine TCR | This paper | This construct was prepared in lab. Detailed construct preparation can be found in Table 1 in main article file. | |
| Recombinant DNA reagent | anti TRP1 murine TCR | This paper | This construct was prepared in lab. Detailed construct preparation can be found in Table 1 in main article file. | |
| Recombinant DNA reagent | TA99 scFv – OVA257-264 – G4S linker - OVA323-339 | This paper | This construct was prepared in lab using linear DNA ordered from (IDT). Detailed construct preparation can be found in Table 1 in main article file. | |
| Recombinant DNA reagent | OVA257-264 – G4S linker - OVA323-339 | This paper | This construct was prepared in lab. Detailed construct preparation can be found in Table 1 in main article file. | |
| Commercial assay, kit | Mouse IgG ELISA Kit | Bethyl | Catalog # E99-131 | |
| Commercial assay, kit | Mouse IgM ELISA Kit | Bethyl | Catalog # E99-101 | |
| Commercial assay, kit | Human Granzyme B ELISA Kit | Abcam | ab235635 | |
| Commercial assay, kit | Griess Reagent System | Promega | Catalog: G2930 | |
| Chemical compound, drug | Trastuzumab | Roche | INF/INJ-HER-2021 02–0 | |
| Chemical compound, drug | Cetuximab | Merck | Erbitux 5 mg/mL solution for infusion | |
| Chemical compound, drug | Rituximab | Genentech | Rituxan | |
| Other | FITC Annexin V | Biolgend | Cat: 640905 | 5 µl in 100 µl volume |
| Other | Propidium iodide (PI) | Biolegend | Cat: 421301 | 0.5 mg/ml |





